ÃÆà ½Ãâñ Globin Hemoglobinopathy as A Case Study: Mutants and Prototype Mutants: Are They Absent ÃÆà ½Ãâñ0, Reduced ÃÆà ½Ãâñ+ or Recombinant?
Al-Fattah Amara AA
DOI10.21767/2573-5365.100033
Amro Abd Al Fattah Amara*
Department of Protein Research, Genetic Engineering and Biotechnology Research Institute, Scientific Publishing, City for Scientific Research and Technological Applications, Universities and Research Center District, New Borg El-Arab, Alexandria, Egypt
- *Corresponding Author:
- Al-Fattah Amara AA
Head of the Protein Research Department, Genetic Engineering and Biotechnology Research Institute, Head of the office of the Scientific Publishing, City for Scientific Research and Technological Applications, Universities and Research Center district, New Borg El-Arab, Alexandria, Egypt.
Tel: +203-4593422
Fax: +203-4593497
E-mail: amroamara@web.de;amroamara@protonmail.fc
Received Date: May 05, 2017; Accepted Date: June 29, 2017; Published Date: June 30, 2017
Citation: Amara AAAF (2017) α Globin Hemoglobinopathy as A Case Study: Mutants and Prototype Mutants: Are They Absent α0, Reduced α+ or Recombinant? Cell Mol Med. Vol. 3 No. 2:10
Abstract
Recently the existence of prototype mutants in β globin genes was proved. In this study a further and similar investigation was performed. The investigation concerns the Homo sapiens α globin. The prototype mutants were screened in the α globin DNA sequences. The results proved their existence. A selected α globin fragment contains the prototype mutant was used for searching both of the nucleotide and protein databases. The used fragment consists of nine nucleotides GGC AAG GTT and contain two silent mutants with the following amino acids sequences GKV. Searching the Blast protein database for the existence of correct mutants using the V00493.7 Homo sapiens 27 amino acids fragment was led to find 50 different sequences. Interestingly the protein sequences show similarity with marsupials, even-toed ungulates, odd-toad ungulates, placental, bats, rodents, carnivores, whales and dolphins; primates, insectivores, rabbits and hares. Species responsible for human bit or blood transfer are located and suggest gene recombination between different species occure through blood transfer. The restriction map for the 81 nucleotides for four nucleotide sequences (one prototype mutant, two correct mutants, and one normal) proved change in the restriction enzymes profile of the α globin sequences. That might be a proof for the existence of reduced α+ globin (also in case of β+ globin) macromolecules. The protein modelling in the selected different fragments show differences in the 3D structure and in their hydrophobicity. The unexpected exo-sources for the α globin put another load on the possibility of the haemoglobin illness solving. However, avoiding marriage from relatives, and from the same ethnic groups might be the best choice. Additionally avoiding blood contact with animals have compatible mutants is recommended. Further investigations are in need to prove/disprove the role of the gene recombination between species through blood in the transfer of the α globin mutants or even other ones.
Keywords
α Globin prototype; Gene recombination; Nucleotides database; Protein database
Introduction
The quaternary structure of the hemoglobin macromolecules consists of four globin polypeptide chains. Like any of the human genes some variant of the globin molecules are existed which controlled by basic genetic rolls. Hemoglobin diseases are a global disease. WHO estimated that 5% of the world populations have hemoglobin disorders [1].
Hemoglobin is the oxygen carrier tetrameric molecule and can be found in vertebrate red blood cells, in some invertebrates and in the root nodules of legumes [1-4]. Livingstone in 1989 described that the disease is concentrated in some parts of the world particularly where the malaria is existed [5] and/or where excessive marriage from relatives over generations is existed as suggested by Amara in 2013 and 2015 [6,7]. Both of the described reasons are responsible for the image of the disease in the Mediterranean region and West Africans [5,7,8].
The hemoglobin molecules are highly sensitive to the O2/ CO2 exchange. The sensitivity level could be in range can find differences between O2 and CO2 molecules. The folded hemoglobin macromolecules are able to differentiate between O2 and CO2. Its four-subunit composition of the normal adult hemoglobin (HbA) are α2β2. The other variants collectively can be found in the fetal hemoglobin (HbF:α2γ2), sickle cell hemoglobin (HbS:α2S2), and a minor adult hemoglobin (HbA2:α2δ2). Amara in 2015 suggested that prototype mutant in β globin could be a hidden source or sleeping mutants that could be converted to complete mutants (in the future). Accumulating, different types of prototype mutants might be avoided if detected early by avoiding marriage between individuals harboring the responsible genes (from each other) [7]. Even that the primary structures of the β, γ, and δ chains of human hemoglobin are highly conserved, human have stages of modifications concerning the functional hemoglobin. In general; fetus initially synthesizes a ζ2ε2 tetramer. After the first trimester, ζ and γ subunits will be replaced by α and ε subunits. While synthesis of β subunits begins in the third trimester, β subunits do not completely replace γ subunits to yield adult HbA (α2β2) until some weeks postpartum. For more details refer to Harper’s Illustrated Biochemistry in 2003 [9].
Over 800 known mutant of the human hemoglobins are both extremely rare and benign, presenting no clinical abnormalities. Hemoglobinopathy is a term given for the mutants that compromise biologic function [9,10]. It is still not fully proved if the subunit is completely absent (α0 or β0) or whether its synthesis is reduced (reduced α+ or β+ ) [9,11-13].
Like any genes hemoglobin genes governed by the same genetic and molecular biology roles. Gillbert wrote about the gene recombination in the hemoglobin during the differentiation “If recombination occurs between two regions of homology that are in different genes (unaligned recombination or unequal crossing over), individual genes can be duplicated or lost in the resulting daughter DNA. A good example is the globin gene family. If recombination occurs between two similar (but not identical) genes, the resulting DNA will be rearranged so that one progeny is a gene or two short while the other offspring has a few too many. Again, no DNA has actually been lost; it’s just been redistributed between offspring” [14]. In the adage of the molecular biology and the genetic engineering, one should not kick out the basic genetic roles which might introduce simpler solutions [6].
This study was started with a single aim to prove only the presence of prototype mutants in the α globin such as that proved by Amara in 2015 [7] in case of β globin genes, however the findings extend that aim and strong evidences about gene recombination between species was found.
Materials and methods
4.1 The Homo sapiens α globin nucleotides sequences collection
Human α globin DNA sequence (AF349571.1 Homo sapiens haemoglobin alpha-1 globin chain (HBA1) mRNA, complete cds) was the start point of this study. For more details about the source gene refer to the information provided in the AF349571.1 document. The sequence was obtained from the www.ncbi.nlm.nih.gov nucleotides database and has transferred to the nucleotide blast search. These nucleotide database search nucleotides using a nucleotide query (Blast.ncbi.nlm.nih.gov/ Blast.cgi) [15].
>AF349571.1
cccacagactcagagagaacccaccatggtgctgtctcctgacgacaagaccaacgt caaggccgcctggggtaaggtcggcgcgcacgctggcgagtatggtgcggaggccctg gagaggatgttcctgtccttccccaccaccaagacctacttcccgcacttcgacctgagc cacggctctgcccaggttaagggccacggcaagaaggtggccgacgcgctgaccaacg ccgtggcgcacgtggacgacatgcccaacgcgctgtccgccctgagcgacctgcac gcgcacaagcttcgggtggacccggtcaacttcaagctcctaagccactgcctgctgg tgaccctggccgcccacctccccgccgagttcacccctgcggtgcacgcctccctgga caagttcctggcttctgtgagcaccgtgctgacctccaaataccgttaagctggagcctc ggtggccatgcttcttgcccctttg
The nucleotides sequences adjustment
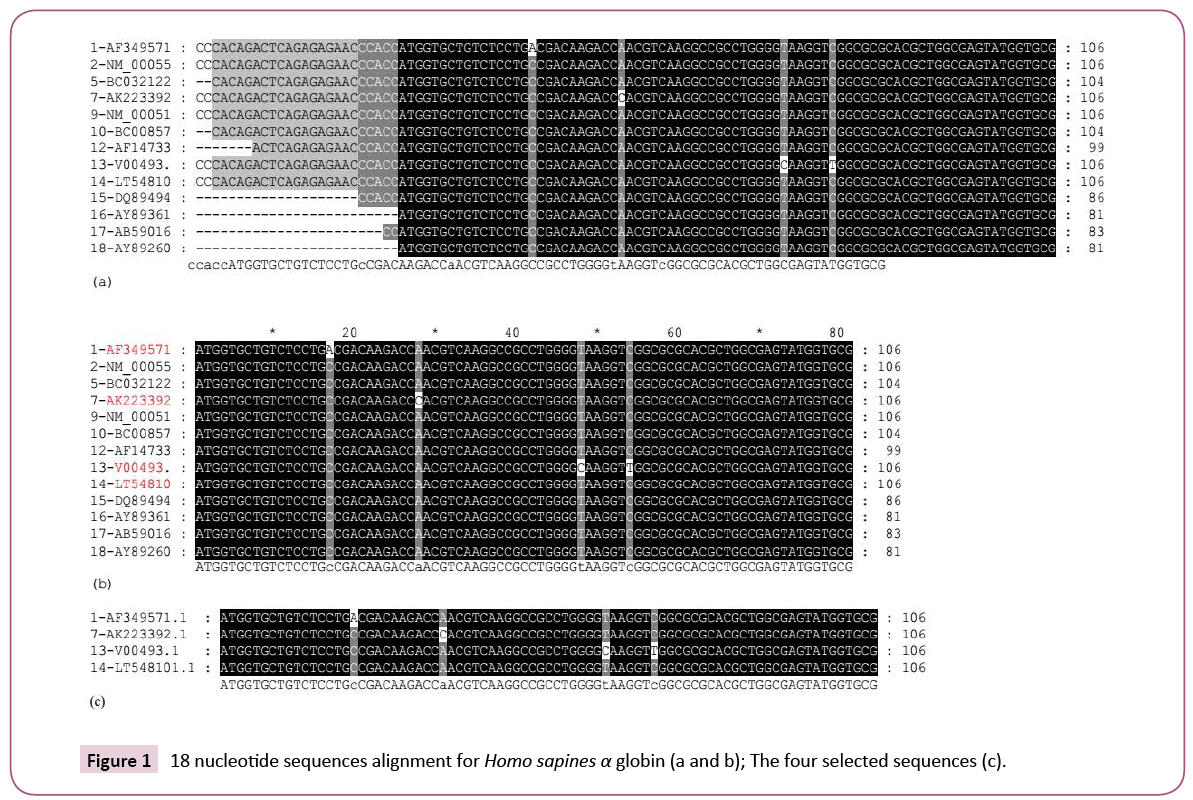
After the search was completed 100 nucleotide sequences were obtained. The non-human nucleotide sequences were removed and only 21 human α globin nucleotide sequences were selected. The sequences were saved in one file as FASTA format. Aiming to exclude similar and repeated names sequences the "Data Analysis in Molecular Biology and Evolution” DAMBE ver 6.4. 58. software was used. Further four similar sequences were removed, and a sum of 18 sequences were kept for further investigation.
Nucleotides sequences alignment
The 18 obtained sequences were aligned using Clustal Xver. 2.1 [16] BioEdit ver 7.2.3. and the sequences were investigated on the GeneDoc 2.7. [17]. Four nucleotide sequences were selected, three contain mutants and one is normal.
Identifying the prototype mutants and select the targeted fragment
Manual selection for the silent mutant (prototype mutant) was performed as described by Amara in 2015 [7]. Four nucleotide sequences were selected. Two sequences, each contain a single correct mutant (single amino acid change) and one sequence contain silent two mutants, and one is normal. The nucleotide fragments which contain the specified mutants (81 nucleotides) were specified and selected for further investigations.
Restriction map
The four nucleotide sequence for the used fragments (81 nucleotides) which represent the AF349571.1; AK223392.1; LT548101.1 and V00493.1 were used against the restriction enzyme database in each of Clone Manager version 4.01 software [18], pDRAW32.10 software [19] and Serial Cloner software 2.6.1. [20]. In each program each sequence was put in single txt file. In clone 4 and Serial Cloner software 2.6.1. the nucleotide sequence was adjusted to FASTA format. In pDRAW32.10 software the nucleotide sequence was adjusted without name as a text file. The restriction map for each sequence using each software was performed. The results were obtained and saved as image in case of the pDRAW32.10 software and Serial Cloner software 2.6.1 or in table as in Clone Manager version 4.01 software.
Tracing the different restriction maps
The restriction maps were screened manually for the existence of different restriction sites.
Screening the blast nucleotide database for similar mutants
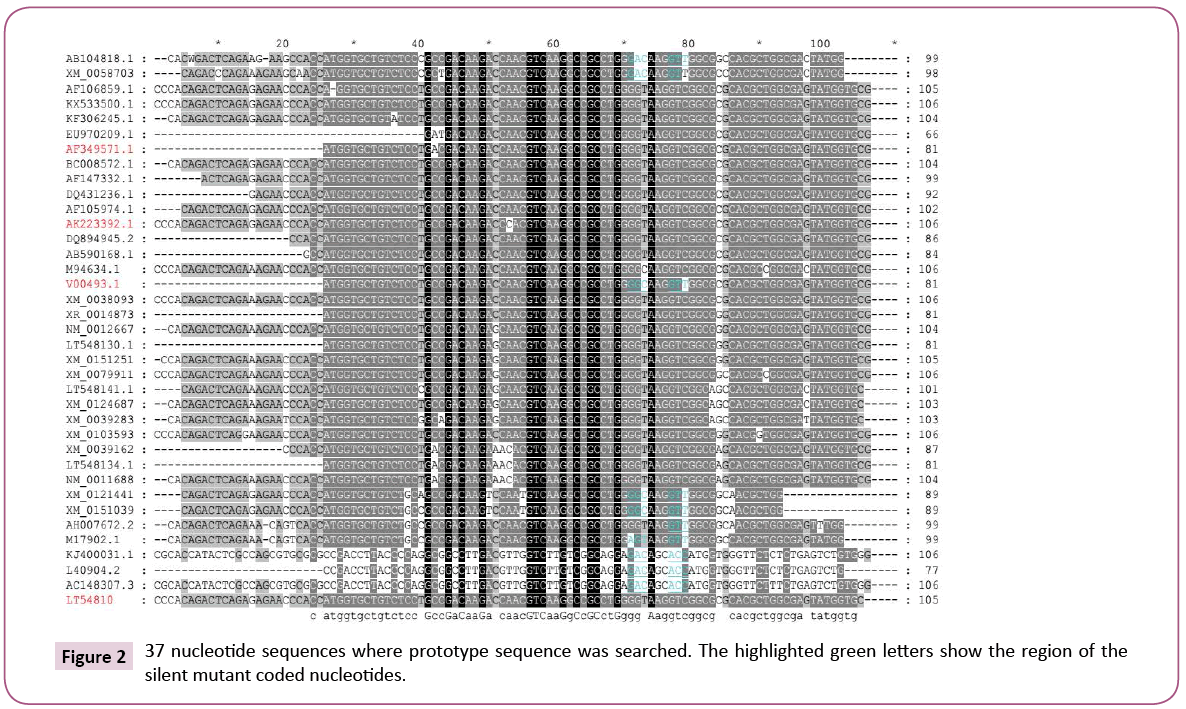
The nucleotide database was screened for the existence of similar mutants using nucleotide query to prove their possible existence. The sequence which specified to contain prototype mutant was used (V00493.1). The obtained data were adjusted to remove any duplicated sequences or similar names (as above) using DAMBE ver 6.4.58 software. Then the rest were aligned using BioEdit ver 7.2.3. software and the alignment was represented using the GeneDoc 2.7. software. The similar mutants were given different colour (green) by manual highlighting.
Nucleotides translation
The selected nucleotide fragment sequences (81 nucleotides) were translated to 27 amino acids using Blastx (search protein database using a translated nucleotide query) and were rechecked using the translation option in BioEdit software ve 7.2.3 software [21].
Screening for correct mutants using 27 amino acids sequence
The following nucleotide sequences (under line) which has prototype mutant in the original nucleotide sequence was translated.
“(V00493.1 Homosapiens messenger mRNA for hemoglobin alpha chain: CCCACAGACTC A G A G A G A A C C C A C C A T G G T G C T G T C T C C T G C C G A CAAGACCAACGTCAAGGCCGCCTGGGGCAAGGTTGGCGCGCAC GCTGGCGAGTATGGTGCGGAGGCCCTGGAGAGGATGTTCCT GTCCTTCCCCACCACCAAGACCTACTTCCCGCACTTCGACCT GAGCCACGGCTCTGCCCAGGTTAAGGGCCACGGCAAGAAG GTGGCCGACGCGCTGACCAACGCCGTGGCGCACGTGGAC GACATGCCCAACGCGCTGTCCGCCCTGAGCGACCTGCACGC GCACAAGCTTCGGGTGGACCCGGTCAACTTCAAGCTCCTA AGCCACTGCCTGCTGGTGACCCTGGCCGCCCACCTCCCCGCCGA GTTCACCCCTGCGGTGCACGCCTCCCTGGACAAGTTCCTGGCTTC TGTGAGCACCGTGCTGACCTCCAAATACCGTTAAGCTGGAGCCTC GGTAGCAGTTCCTCC)” The translated amino acids are MVLSPADKTNVKAAWGKVGAHAGEYGA were used for screening Swissport Database to find any change in the two targeted amino acids (G and V).
1 ATG GTG CTG TCT CCT GCC GAC AAG ACC AAC GTC AAG GC C GCC TGG GGC AAG GTT GGC GCG CAC GCT GGC GAG TAT G GT GCG 81
1 Met Val Leu Ser Pro Ala Asp Lys Thr Asn Val Lys Ala Ala Trp Gly Lys Val Gly Ala His Ala Gly Glu Tyr Gly Ala 27
ATGGTGCTGTCTCCTGCCGACAAGACCAACGTCAAGGCCGCCTG GGGCAAGGTTGGCGCGCACGCTGGCGAGTATGGTGCGMVLSP ADKTNVKAAWGKVGAHAGEYGA
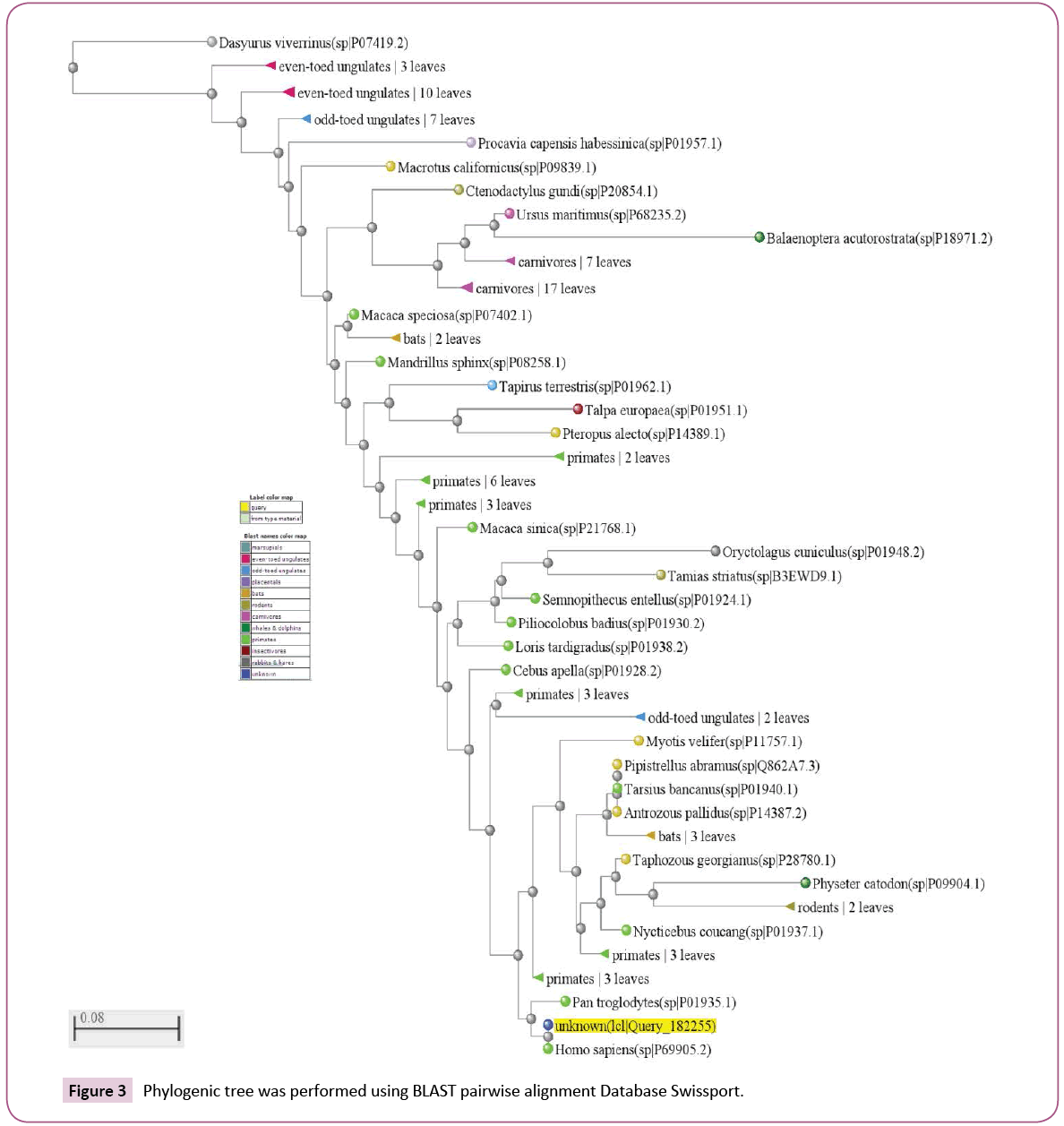
The number of searched sequences output was adjusted to give 100 results. The obtained sequences were alignment using Swissport Database search tools and the phylogenic tree was performed (Figure 1). Additionally the tree was visualized offline as circular tree using Tree Figure Drawing Tool Version 1.4.2 software after adjusting the species names. The tree obtained from the Swissport Database summarised the species based on their types. The alignment result was saved for offline manipulation.
Refining the protein sequences
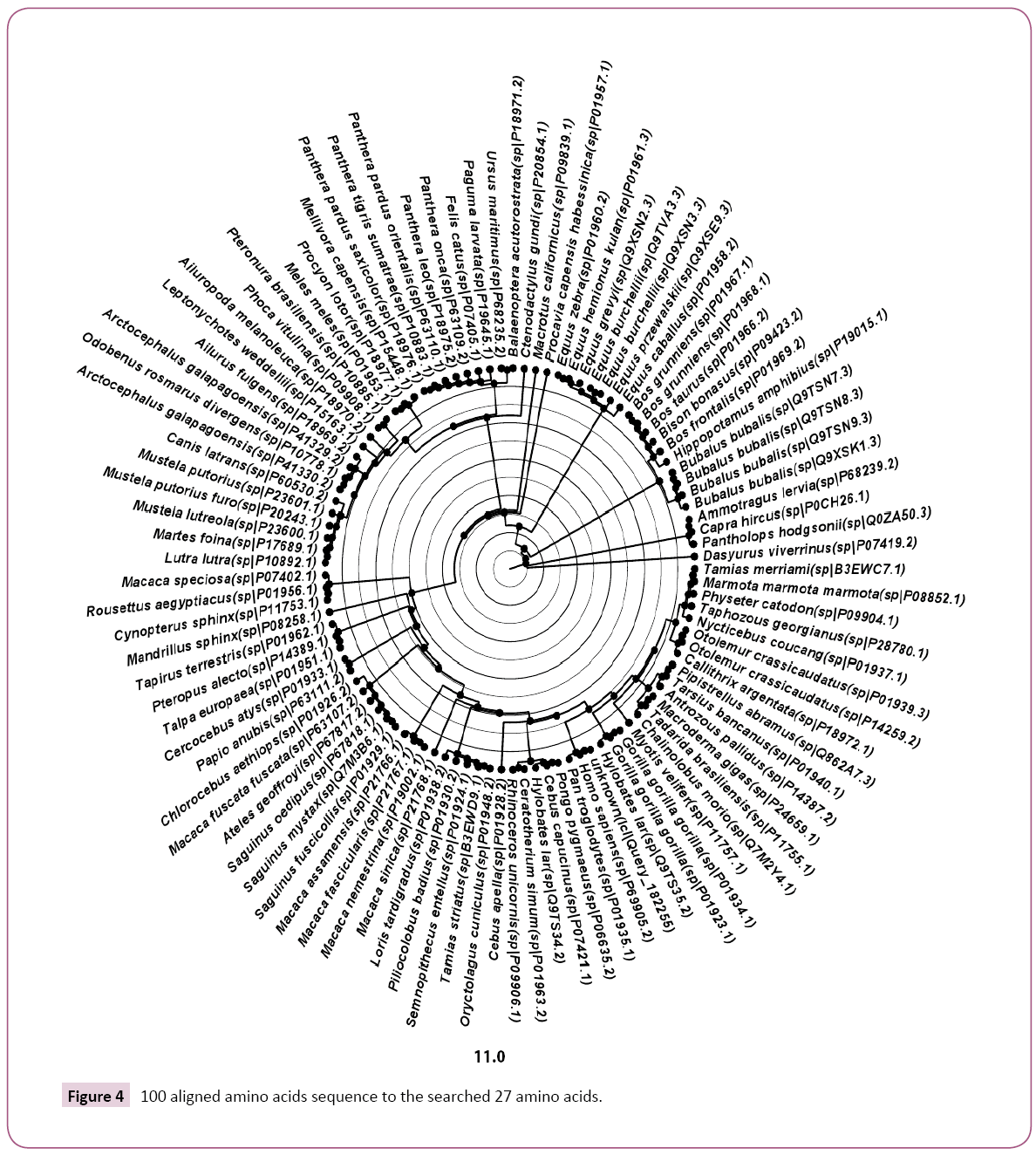
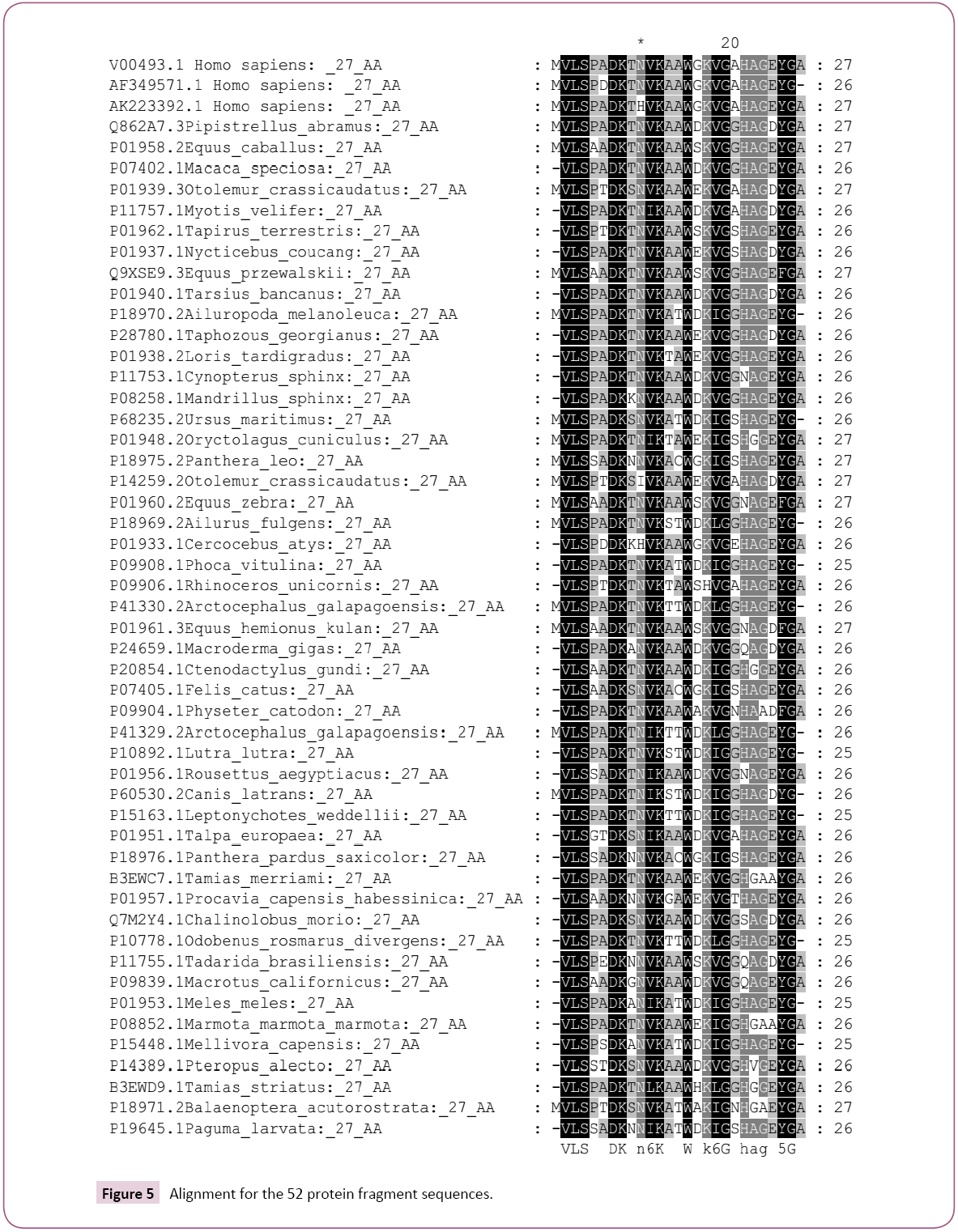
The obtained 100 protein sequences were refined using DAMBE ver 6.4.58 software to remove the repeated and similar names and sequences. Then manual remove for the sequences which have no change in the targeted GKV region was performed. At the end out of 100 protein sequences only 50 were obtained. Manually additional two sequences were loaded (AF349571.1 Homo sapiens and AK223392.1 Homo sapiens). Full alignment for the 52 sequences was performed and the phylogenic tree was obtained using Mega 6 software (Figures 2 and 3).
Phylogenic tree generation and visualization
Phylogenic tree and the alignment for selected proteins and nucleotides of the α globin were generated using different software (few only were represented). One phylogenic tree was obtained from the online BLAST pairwise alignments option in Swissport Database. The offline used software for trees generation were, BioEdit ver 7.2.3., Mega 6 software, DAMBE ver 6.4.58 software and Clustalx ver 2.1. Tree Figure Drawing Tool Version 1.4.2 was used for visualizing each of the obtained trees [22]. Mega 6 software was used to give statistical report
Protein models generation
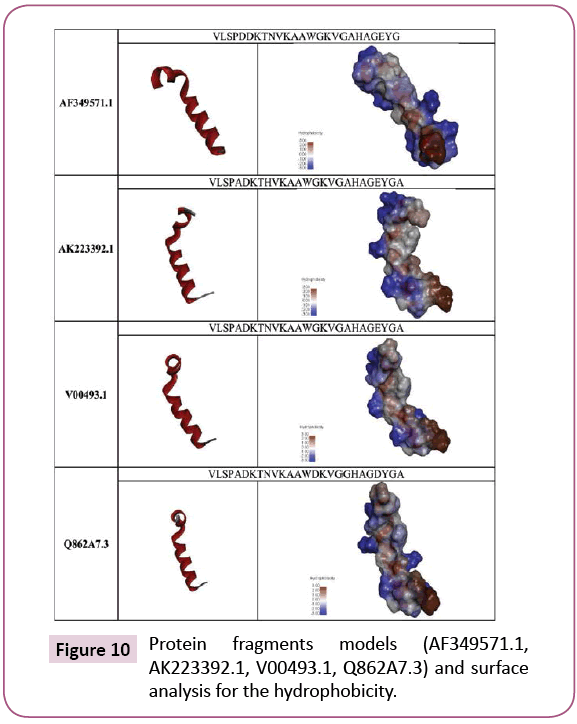
Model for each of AF349571.1; AK223392.1; V00493.1 and Q862A7.3 α globin 27 amino acids fragments were generated. The Hemoglobin pdb file (P69905) was used to obtain single α globin macromolecule by the aid of the software Discovery Studio 4.1. Client 4.1.0.14169 (Accelrys software Inc.) software. The α globin used protein sequence was as the follow: The SEQUENCE 142 AA; 15258 MW; 15E13666573BBBAE CRC64; MVLSPADKTNVKAAWGKVGAHAGEYGAEALERMFLSFPTTKTYFP HFDLSHGSAQVKGHGKKVADALTNAVAHVDDMPNALSALSDLHA HKLRVDPVNFKLLSHCLLVTLAAHLPAEFTPAVHASLDKFLASVSTV LTSKYR.
The software Modeller v 9.8 was used for generating model for each protein fragment (27 amino acids) [23]. For better 3D structure, the background of the images have been converted to white and the 3D image have been adjusted and saved [24] (Figures 4 and 5). The protein fragment was represented as ribbon and the surface structure of the hydrophobicity of the fragment was performed using the Discovery Studio 4.1. Client 4.1.0.14169 (Accelrys software Inc.) software. Only the 3D structure and the hydrophobicity were used to visualize the protein 3D differences due to the different amino acid constituents in the selected fragment.
Results and Discussion
Hemoglobin diseases are impotent due to their health and economic side effect. Hemoglobin diseases are genetically based. They are hard to be repaired, at least in the few coming years or till significant progress happened in the gene therapy [6]. Each somatic cell has the defect gene. β globin macromolecules proved to have prototypes mutants in their nucleotide sequences [7]. This study represents an investigation concerning α globin. The study aims to prove or disprove that α globin macromolecules have prototypes mutants. The investigation strategy is based on specifying some mutants on the α globin DNA then translating them. Finding silent mutation is the start point to find others converted to complete mutants. That was performed by manual tracing the prototype mutants obtained from the Homo sapiens nucleotide sequence alignment as in Figure 6.
The study was started by searching the blast protein database against a complete α globin sequence for 100 nucleotide sequences. The result contains Homo sapiens and other species genes. 21 Homo sapiens genes only were located and were selected (Figure 6). The Homo sapiens genes were adjusted to FASTA format and were investigated using the DAMBE software. The software enables removing of similar sequences and detecting sequences with similar names. Three similar sequences were removed. The 18 different sequences were subjected to alignment (Figure 7). The result proves the existence of different mutants in the different sequences. Four sequences were selected. Two sequences each contain a single nucleotide mutant; one contains two nucleotide mutants and one did not contain mutants (in the selected part). The Homo sapiens selected nucleotides sequences were subjected to logical analysis for the mutant and the prototype mutants as in (Figure 6).
Four sequences were selected represent each of
1. AF349571.1 Homo sapiens haemoglobin alpha-1 globin chain (HBA1) mRNA, complete cds
2. AK223392.1 Homo sapiens mRNA for alpha 2 globin variants, clone: FCC107C11
3. V00493.1 Homo sapiens messenger mRNA for haemoglobin alpha chain
4. LT548101.1 TPA_inf: Homo sapiens GLNC1 gene for globin C1
Searching for the existence of similar prototype mutants in the nucleotides blast database that prove their existence as in Figure 7. The nucleotide sequences were translated to their amino acids each of AF349571.1 and AK223392.1 show correct single amino acid change due to a single nucleotide change. Sequence V00493.1 show two nucleotides changes but no amino acid change (silent mutants) or as suggested by Amara in 2015 prototype mutants (Table 1) and one is normal.
| >1-AF349571.1 (complete) CCCACAGACTCAGAGAGAACCCACCATGGTGCTGTCTCCTGACGACAAGACCAACGTCAAGGCCGCCTGGGGTAAGGTCG GCGCGCACGCTGGCGAGTATGGTGCGGAGGCCCTGGAGAGGATGITCCTGTCCITCCCCACCACCAAGACCTACTTCCCG CAC1TCGACCTGAGCCACGGCTCTGCCCAGG1TAAGGGCCACGGCAAGAAGGTGGCCGACGCGCTGACCAACGCCGTGGC GCACGTGGACGACATGCCCAACGCGCTGTCCGCCCTGAGCGACCTGCACGCGCACAAGCTTCGGGTGGACCCGGTCAACT TCAAGCTCCTAAGCCACTGCCTGCTGGTGACCCTGGCCGCCCACCTCCCCGCCGAG1TCACCCCTGCGGTGCACGCCTCC CTGGACAAGITCCTGGCITCTGTGAGCACCGTGCTGACCTCCAAATACCGTTAAGCTGGAGCCTCGGTGGCCATGC1TCT TGCCCCT1TGG |
| >1-AF349571 (81 nucleotides and 27 amino acids) 1 ATC CIO CSC 1C2 CC2 CAC CAC AAC ACC AAC CIC AkOCCCGEC !GCGC! MC CSC GCE CCC CAC Ca GCE CAC 2AI 0= CCC SI 1 Mat Val Le. See Pee Asp Asp Lys She Ask Val Lys Ala Ala Sep Cly Lys Val Cly Ala Nis Ala Cly Cl. IyeCly Ala 27 ATGGTGCTGTCTCCTGACGACAAGACCAACGTCAAGGCCGCCTGGGGTAAGGTCGGCGCGCACGCTGGCGAGTATGGTGCG MVLSPDDKTNVKAAWGKVGAHAGEYG |
| >7-AK223392.1 (complete)GCGCGCACGCTGGCGAGTATGGTGCGGAGGCCCTGGAGAGGATGITCCTGTCC1TCCCCACCACCAAGACCTACTTCCCG CAC1TCGACCTGAGCCACGGCTCTGCCCAGG1TAAGGGCCACGGCAAGAAGGTGGCCGACGCGCTGACCAACGCCGTGGC GCACGTGGACGACATGCCCAACGCGCTGTCCGCCCTGAGCGACCTGCACGCGCACAAGCTTCGGGTGGACCCGGTCAACT TCAAGCTCCTAAGCCACTGCCTGCTGGTGACCCTGGCCGCCCACCTCCCCGCCGAG1TCACCCCTGCGGTGCACGCCTCC CTGGACAAGITCCTGGCITCTGTGAGCACCGTGCTGACCTCCAAATACCGTTAAGCTGGAGCCTCGGTGGCCATGC1TCT TGCCCCTT |
| >7 -AK223392.1 1 AIC CSC CSC 2C2 CC2 GCC CAC AAC ACC CAC CSC AAC CCC CCC DSC 0 AAC CSC CrX GEC VC CC2 CrX CAC nt CO[ CCC 11 1 Mat Val La. Sae PCO Ala A.) Lya She N“ ValLya Ala Ala 20 Cly Lyn Val Cly Ala NIa Ala Cly CI. IycCly Ala 27 ATGGTGCTGTCTCCTGCCGACAAGACCCACGTCAAGGCCGCCTGGGGTAAGGTCGGCGCGCACGCTGGCGAGTATGGTGCG MVLSPADKTHVKAAWGKVGAHAGEYGA |
| >13-V00493.1 (complete) CCCACAGACTCAGAGAGAACCCACCATGGTGCTGTCTCCTGCCGACAAGACCAACGTCAAGGCCGCCTGGGGCAAGGTTG GCGCGCACGCTGGCGAGTATGGTGCGGAGGCCCTGGAGAGGATGTTCCTGTCCTTCCCCACCACCAAGACCTACTTCCCG CACTTCGACCTGAGCCACGGCTCTGCCCAGGTTAAGGGCCACGGCAAGAAGGTGGCCGACGCGCTGACCAACGCCGTGGC GCACGTGGACGACATGCCCAACGCGCTGTCCGCCCTGAGCGACCTGCACGCGCACAAGCTTCGGGTGGACCCGGTCAACT TCAAGCTCCTAAGCCACTGCCTGCTGGTGACCCTGGCCGCCCACCTCCCCGCCGAGTTCACCCCTGCGGTGCACGCCTCC CTGGACAAGTTCCTGGCTTCTGTGAGCACCGTGCTGACCTCCAAATACCGTTAAGCTGGAGCCTCGGT |
| >13-V00493.1 1 ATG GTG CTG TCT CCT GCC GAC AAG ACC AAC GTC AAG GCC GCC TGG GGC AAG GTT GGC GCG CAC GCT GGC GAG TAT GGT GCG 81 1 Met Val Leu Ser Pro Ala Asp Lys ThrAsn Val Lys Ala AlaTrpGly Lys Val Gly Ala His Ala GlyGlu Tyr Gly Ala 27 ATGGTGCTGTCTCCTGCCGACAAGACCAACGTCAAGGCCGCCTGGGGCAAGGTTGGCGCGCACGCTGGCGAGTATGGTGCG MVLSPADKTNVKAAWGKVGAHAGEYGA [Prototype mutant; the amino acids still normai] |
| >14-LT54810.1 (complete) CCCACAGACTCAGAGAGAACCCACCATGGTGCTGTCTCCTGCCGACAAGACCAACGTCAAGGCCGCCTGGGGTAAGGTCG GCGCGCACGCTGGCGAGTATGGTGCGGAGGCCCTGGAGAGGATGTTCCTGTCCTTCCCCACCACCAAGACCTACTTCCCG CACTTCGACCTGAGCCACGGCTCTGCCCAGGTTAAGGGCCACGGCAAGAAGGTGGCCGACGCGCTGACCAACGCCGTGGC GCACGTGGACGACATGCCCAACGCGCTGTCCGCCCTGAGCGACCTGCACGCGCACAAGCTTCGGGTGGACCCGGTCAACT TCAAGCTCCTAAGCCACTGCCTGCTGGTGACCCTGGCCGCCCACCTCCCCGCCGAGTTCACCCCTGCGGTGCACGCCTCC CTGGACAAGTTCCTGGCTTCTGTGAGCACCGTGCTGACCTCCAAATACCGTTAA |
| >14-LT54810.1 1 ATG GTG CTG TCT CCT GCC GAC AAG ACC AAC GTC AAG GCC GCC TGG GGT AAG GTC GGC GCG CAC GCT GGC GAG TAT GGT GCG 81 1 Met Val Leu Ser Pro Ala Asp Lys ThrAsn Val Lys Ala AlaTrpGly Lys Val Gly Ala His Ala GlyGlu Tyr Gly Ala 27 ATGGTGCTGTCTCCTGCCGACAAGACCAACGTCAAGGCCGCCTGGGGTAAGGTCGGCGCGCACGCTGGCGAGTATGGTGCG MVLSPADKTNVKAAWGKVGAHAGEYGA |
Table 1: Logical analysis for the existence of prototype mutants.
By taking the protein fragment where the prototype mutants were specified and searching in the protein database for the probable existence of complete mutants, the results proved the existence of complete change in the specified amino acids as in (Figures 1-3,8). That proves the possibility of changing the prototype mutant (silent mutants) to complete correct mutants (amino acids changes). Unexpected result was emerged that the mutants were founds in organisms either able to bit the human or able to transfer blood from other species. That might be open the window for new explanation for the role of the natural gene transfer by insects and animals as in Figure 8. The symptoms of the rabies (the disease transferred by doges) including changing the infected patient behavior. Rabies is an acute viral disease of the nervous system of warm-blooded animals (usually transmitted by the bite of a rabid animal); rabies is fatal if the virus reaches the brain).
The V00493.1 nucleotide sequence contains two nucleotide changes and given no amino acids change which means that these two mutants are silent mutants. This sequence attracts the attention for investigating if those two silent mutants were mutated in any other sequence of the sequences located in the nucleotide database. The nucleotide database was searched using the selected 81 V00493.1 nucleotide sequence and the results proved the existence of similar mutants as in Figure 7. The similar mutants were highlighted by green color (Figure 7). The selected fragment sequences constituent of 81 nucleotides were translated to 27 amino acids using Blastx (search protein database using a translated nucleotide query) and were rechecked using the translation option in BioEdit software version 7.2.3 software. The translated 81 amino acids (V00493.1) protein fragment was used to search similar protein using Swissport database against 100 results. The obtained results prove the existence of correct mutants representing those two suspected amino acids (GKV). As above the obtained 100 sequences were preliminarily evaluated using DAMBE ver 6.4.58 software. The total sequences finally become as 50 sequences and additional two sequences were added manually (AF349571.1 and AK223392.1) as in Figures 2. The first obtained 100 sequences were subjected to phylogenetic tree analysis and proved that species other than the Homo sapiens are represented (Figure 1). Both of the 100 and the 52 sequences which were used either for the tree (Figures 1 and 3) generation or for the alignments prove some facts (Figure 2):
1. Prototype mutants could become full correct mutants
2. There is possibility for gene transfer within species.
3. The α globin genes prototype mutants are more likely to change to full mutants by the activity of different types of genetic modifications including epigenetic, cross-over, recombination, splicing, methylation, blood transfer etc.
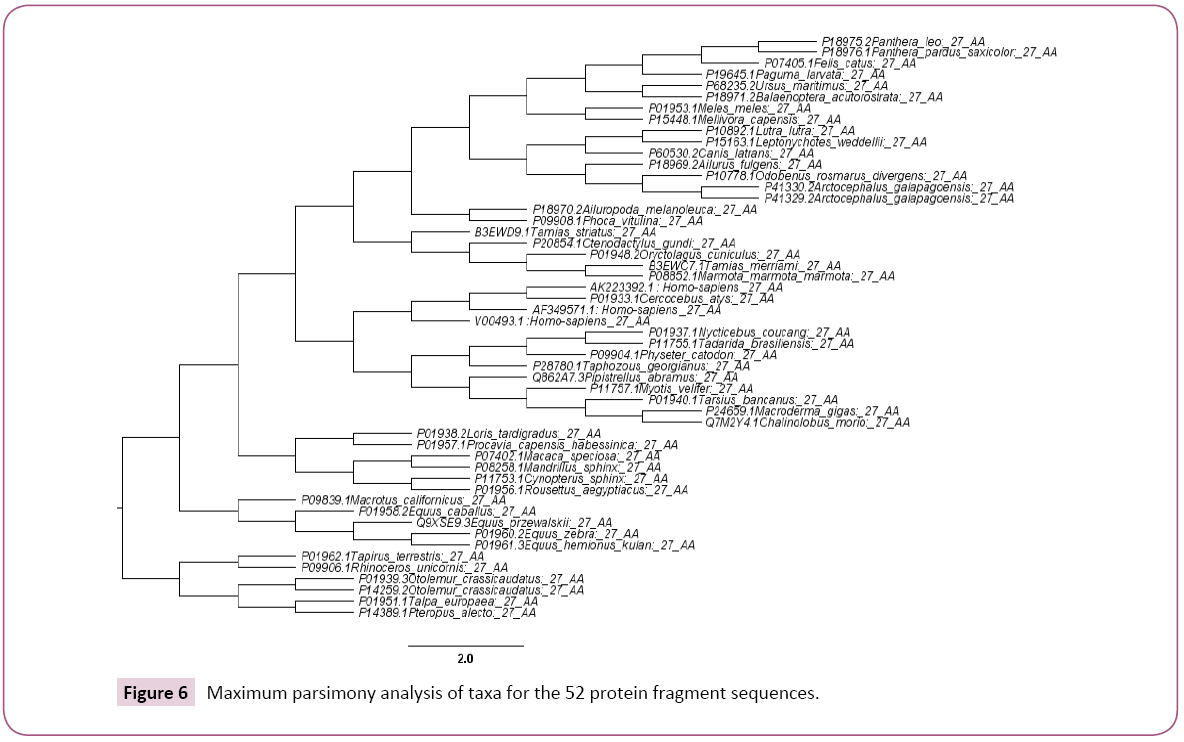
In Figure 3 which represents the phylogenic tree of the 52-selected sequence, the evolutionary history was inferred using the Maximum Parsimony method. The most parsimonious tree with length=94 is shown. The consistency index is 0.457447 (0.451613), the retention index is 0.653061 (0.653061), and the composite index is 0.298741 (0.294931) for all sites and parsimony-informative sites (in parentheses). The MP tree was obtained using the Subtree-Pruning-Regrafting (SPR) algorithm (pg. 126 in ref. [25]) with search level 0 in which the initial trees were obtained by the random addition of sequences (10 replicates). The analysis involved 52 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 25 positions in the final dataset. Evolutionary analyses were conducted in MEGA6 [26].
Even this study proves that more resources for the globin mutation are existed but solutions also could be suggested based on the type and the source of those mutants. For example, the prevention of the existence of heterozygote alleles is so simple, just by totally avoiding the marriage between the offspring of the homozygote patients for several generations [6,7]. As in the folk experiences; for seven-generation [6]. The marriage out of the hemoglobin patients’ carrier will lead to diluting the probability of the existence of the gene generation after generation. Even it might finally lead to the disappear of the disease. The species which showed alignment with the 27 searched amino acids are: V00493.1 Homo sapiens; AF349571.1 Homo sapiens; AK223392.1 Homo sapiens; Q862A7.3 Pipistrellus abramus; P01958.2 Equus caballus; P07402.1 Macaca speciosa; P01939.3 Otolemur crassicaudatus; P11757.1 Myotis velifer; P01962.1 Tapirus terrestris; P01937.1 Nycticebus coucang; Q9XSE9.3 Equus przewalskii; P01940.1 Tarsius bancanus; P18970.2 Ailuropoda melanoleuca; P28780.1 Taphozous georgianus; P01938.2 Loris tardigradus; P11753.1 Cynopterus sphinx; P08258.1 Mandrillus sphinx; P68235.2 Ursus maritimus; P01948.2 Oryctolagus cuniculus; P18975.2 Panthera leo; P14259.2 Otolemur crassicaudatus; P01960.2 Equus zebra; P18969.2 Ailurus fulgens; P01933.1 Cercocebus atys; P09908.1 Phoca vitulina; P09906.1 Rhinoceros unicornis; P41330.2 Arctocephalus galapagoensis; P01961.3 Equus hemionus kulan; P24659.1 Macroderma giga; P20854.1 Ctenodactylus gundi; P07405.1 Felis catus; P09904.1 Physeter catodon; P41329.2 Arctocephalus galapagoensi; P10892.1 Lutra lutra; P01956.1 Rousettus aegyptiacus; P60530.2 Canis latrans; P15163.1 Leptonychotes weddellii; P01951.1 Talpa europaea; P18976.1 Panthera pardus saxicolor; B3EWC7.1 Tamias merriami; P01957.1 Procavia capensis habessinica; Q7M2Y4.1 Chalinolobus morio; P10778.1 Odobenus rosmarus divergens; P11755.1 Tadarida brasiliensis; P09839.1 Macrotus californicus; P01953.1 Meles meles; P08852.1 Marmota marmota marmota; P15448.1 Mellivora capensis; P14389.1 Pteropus alecto; B3EWD9.1 Tamias striatus; P18971.2 Balaenoptera acutorostrata; P19645.1 Paguma larvata; V00493.1 Homo sapiens; AF349571.1 Homo sapiens; AK223392.1 Homo sapiens; Q862A7.3 Pipistrellus-abramus; P01958.2 Equus-caballus; P07402.1 Macaca-speciosa; P01939.3 Otolemur-crassicaudatus; P11757.1 Myotis-velifer; P01962.1 Tapirus-terrestris; P01937.1 Nycticebus-coucang; Q9XSE9.3 Equus-przewalskii; P01940.1 Tarsius-bancanus; P18970.2 Ailuropoda-melanoleuca; P28780.1 Taphozous-georgianus; P01938.2 Loris-tardigradus; P11753.1 Cynopterus-sphinx; P08258.1 Mandrillus-sphinx; P68235.2 Ursus-maritimus; P01948.2 Oryctolagus-cuniculus; P18975.2 Panthera-leo; P14259.2 Otolemur-crassicaudatus; P01960.2 Equus-zebra; P18969.2 Ailurus-fulgens; P01933.1 Cercocebusatys; P09908.1 Phoca-vitulina; P09906.1 Rhinoceros-unicornis; P41330.2 Arctocephalus-galapagoensis; P01961.3 Equushemionus- kulan; P24659.1 Macroderma-giga; P20854.1 Ctenodactylus-gundi; P07405.1 Felis-catus; P09904.1 Physetercatodon; P41329.2 Arctocephalus-galapagoensi; P10892.1 Lutralutra; P01956.1 Rousettus-aegyptiacus; P60530.2 Canis-latrans; P15163.1 Leptonychotes-weddellii; P01951.1 Talpa-europaea; P18976.1 Panthera-pardus-saxicolor; B3EWC7.1 Tamiasmerriami; P01957.1 Procavia-capensis-habessinica; Q7M2Y4.1 Chalinolobus-morio; P10778.1 Odobenus-rosmarus-divergens; P11755.1 Tadarida-brasiliensis; P09839.1 Macrotus-californicus; P01953.1 Meles-meles; P08852.1 Marmota-marmotamarmota; P15448.1 Mellivora-capensis; P14389.1 Pteropusalecto; B3EWD9.1 Tamias-striatus; P18971.2 Balaenopteraacutorostrata and P19645.1 Paguma-larvata.
The restriction maps of the four nucleotide sequences for the used fragments (81 nucleotides) which represent the AF349571.1; AK223392.1; LT548101.1 and V00493.1 were used against the restriction enzyme database in each of Clone Manager version 4.01 software [18], pDRAW32.10 software [19] and Serial Cloner software 2.6.1.[20]. The results were summarized in Figures 4 and 5 and in Table 2. The results show different enzyme profiles. The most interesting result is the appearance of new enzymes that could cut in the nucleotide sequence of V00493.1 sequence as in Figures 4 and 5 and in Table 2. Those new restriction enzymes might suggest the existence of α0 globin or reduced α+ structure. That might happened at the gene level and represented after the protein expression. The Homo sapiens 27 amino acids fragment sequence of AF349571.1, AK223392.1, V00493.1 and Q862A7.3 3D model was built using Modeller v 9.8 software and Discovery Studio 4.1. Client 4.1.0.14169 (Accelrys software Inc.) software. Their 3D structures and their hydrophobicity surface show differences due to their amino acids constituents as in Figures 9 and 10.
| V00493.1 | LT548101.1 | AF349571.1 | AK223392.1 |
|---|---|---|---|
| AccII 1 57 AciI 1 38 AeuI 1 41 AorI 1 41 ApyI 1 41 AtuBI 1 41 BceRI 1 57 BepI 1 57 BsaJI 1 41 BsmAI 1 9 Bsp211I 1 36 BspNI 1 41 BspRI 1 36 BspWI 1 47 BssHII 1 56 BstGII 1 41 BstNI 1 41 BstUI 1 57 BsuRI 1 36 CfoI 2 56, 58 CltI 1 36 CthII 1 41 CviJI 1 36 DsaII 1 36 DsaV 1 41 EcoRII 1 41 FbrI 1 37 Fnu4H 1 37 FnuDI 1 36 FnuDII 1 57 FnuDIII 2 56, 58 HaeIII 1 36 HhaI 2 56, 58 HinPI 2 56, 58 MaeII 1 29 Msp67I 1 41 MvaI 1 41 MvnI 1 57 NgoPII 1 36 PalI 1 36 SciNI 2 56, 58 ScrFI 1 41 SecI 1 41 SfaI 1 36 SsoII 1 41 SuaI 1 36 TaqXI 1 41 ThaI 1 57 ZanI 1 41 |
AccII 1 57 AciI 1 38 AeuI 1 41 AorI 1 41 ApyI 1 41 AtuBI 1 41 BceRI 1 57 BepI 1 57 BsaJI 1 41 BsmAI 1 9 Bsp211I 1 36 BspNI 1 41 BspRI 1 36 BssHII 1 56 BstGII 1 41 BstNI 1 41 BstUI 1 57 BsuRI 1 36 CfoI 2 56, 58 CltI 1 36 CthII 1 41 CviJI 1 36 DsaII 1 36 DsaV 1 41 EcoRII 1 41 FbrI 1 37 Fnu4H 1 37 FnuDI 1 36 FnuDII 1 57 FnuDIII 2 56, 58 HaeIII 1 36 HhaI 2 56, 58 HinPI 2 56, 58 MaeII 1 29 Msp67I 1 41 MvaI 1 41 MvnI 1 57 NgoPII 1 36 PalI 1 36 SciNI 2 56, 58 ScrFI 1 41 SecI 1 41 SfaI 1 36 SsoII 1 41 SuaI 1 36 TaqXI 1 41 ThaI 1 57 ZanI 1 41 |
AccII 1 57 AciI 1 38 AeuI 1 41 AorI 1 41 ApyI 1 41 AtuBI 1 41 BceRI 1 57 BepI 1 57 BsaJI 1 41 BsmAI 1 9 Bsp211I 1 36 BspNI 1 41 BspRI 1 36 BssHII 1 56 BstGII 1 41 BstNI 1 41 BstUI 1 57 BsuRI 1 36 CfoI 2 56, 58 CltI 1 36 CthII 1 41 CviJI 1 36 DsaII 1 36 DsaV 1 41 EcoRII 1 41 FbrI 1 37 Fnu4H 1 37 FnuDI 1 36 FnuDII 1 57 FnuDIII 2 56, 58 HaeIII 1 36 HhaI 2 56, 58 HinPI 2 56, 58 MaeII 1 29 Msp67I 1 41 MvaI 1 41 MvnI 1 57 NgoPII 1 36 PalI 1 36 SciNI 2 56, 58 ScrFI 1 41 SecI 1 41 SfaI 1 36 SsoII 1 41 SuaI 1 36 TaqXI 1 41 ThaI 1 57 ZanI 1 41 |
AccII 1 57 AciI 1 38 AeuI 1 41 AorI 1 41 ApyI 1 41 AtuBI 1 41 BceRI 1 57 BepI 1 57 BsaJI 1 41 BsiYI 1 27 BslI 1 27 BsmAI 1 9 Bsp211I 1 36 BspNI 1 41 BspRI 1 36 BssHII 1 56 BstGII 1 41 BstNI 1 41 BstUI 1 57 BsuRI 1 36 CfoI 2 56, 58 CltI 1 36 CthII 1 41 CviJI 1 36 DsaII 1 36 DsaV 1 41 EcoRII 1 41 FbrI 1 37 Fnu4H 1 37 FnuDI 1 36 FnuDII 1 57 FnuDIII 2 56, 58 HaeIII 1 36 HhaI 2 56, 58 HinPI 2 56, 58 MaeII 1 29 Msp67I 1 41 MvaI 1 41 MvnI 1 57 NgoPII 1 36 PalI 1 36 SciNI 2 56, 58 ScrFI 1 41 SecI 1 41 SfaI 1 36 SsoII 1 41 SuaI 1 36 TaqXI 1 41 ThaI 1 57 ZanI 1 41 |
Table 2: Restriction map performed by Clone 4 software.
Conclusion
In conclusion, this study proves the existence of the prototype mutants in the Homo sapiens and other species of the α globin. The prototype mutants present in full mutants.
The protein structure is modified due to such mutants as well as the restriction map. The existence of the homology between Homo sapiens and the other species particularly the blood transferring ones suggest gene transfer and recombination between species. The facts and the suggestions and the results obtained from this preliminary study need more prove at the cell level to prove or disprove their correction however, they highlight the power of the bioinformatics as science to touch some scientific facts by linking finding and the probability to things one could find in our life but not proved scientifically yet.
Conflict of Interest
The authors have no conflict of interests to declare.
Financial Disclosure
Institutional support
Acknowledgement
The author acknowledges the City of Scientific Research and Technological Applications for supporting this work.
References
- Weatherall DJ, Clegg JB, Higgs DR, Wood WG (1989) The hemoglobinopathies in: Scriver CR, Beaudet AL, Sly WS, Valle D (eds.) The metabolic basis of inherited disease, (6th edn) MaGraw-Hill, New York, USA.
- Aguileta G, Bielawski JP, Yang Z (2006) Evolutionary rate variation among vertebrate beta globin genes: Implications for dating gene family duplication events. Gene 380: 21-29.
- McKusick VA (1990) Mendelian inheritance in man (9th edn) Johns Hopkins University Press, Baltimore, USA.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876-4882.
- Livingstone FB (1989) Simulation of the diffusion of the beta-globin variants in the Old World. Hum Biol 61: 297-309.
- Amara AA (2013) The inevitability of balanced lives: Genes – foods- action-interactions. IIOBJ 4: 1-27.
- Amara AA (2015) The need for early detection of the prototype mutants: Sickle Cell Anemia as a case study. J Proteomics Bioinform S8: 6.
- Kumar R, Sagar C, Sharma D, Kishor P (2015) β-globin genes: Mutation hotspots in the global thalassemia belt. Hemoglobin 39: 1-8.
- Rodwell VW, Kennelly PJ (2003) Chapter 6: Proteins: Myoglobin andhemoglobin In Harper's. In Harper’s Illustrated Biochemistry by Murray RK, Granner DK, Mayes PA, Rodwell VW. Lange Medical Books/McGraw-Hill.
- Discovery Studio Visualizer (2017) V-. Accelrys Software Inc.
- Bunn HF (1997) Pathogenesis and treatment of sickle cell disease. N Engl J Med 337: 762-769.
- Mario N, Baudin B, Giboudeau J (1998) Qualitative and quantitative analysis of hemoglobin variants by capillary isoelectric focusing. J Chromatogr B Biomed Sci Appl 706: 123-129.
- Reed W, Vichinsky EP (1998) New considerations in the treatment of sickle cell disease. Annu Rev Med 49: 461-474.
- Gilbert HF (2000) Chapter 5 Expression of genetic information In Basic concepts in biochemistry a student's survival guide. McGraw-Hill.
- Madden T (2002) Oct 9 (Updated 2003 Aug 13) The BLAST Sequence analysis tool. In: McEntyre J, Ostell J(eds). The NCBI Handbook [Internet]. Bethesda (MD): National Center for Biotechnology Information (US) Chapter 16.
- Thompson MW, McInnes RR, Willard HF (1991) Chapter 11 "The hemoglobinopathies models of molecular disease" In "Genetics in medicine". USA. W. B. Saunders Company. Harcourt Brace Jovanovich, Inc.
- Nicholas KB, Nicholas HB (1997) Jr. GeneDoc: A tool for editing and annotating multiple sequence alignments. Distributed by the author.
- Clone Manager version. (2016) Scientific and Education Software.
- pDRAW32.10.(2009) DNA cloning, sequence analysis and plasmid.
- Perez F. (2004-2013) Serial Cloner. Serial Cloner.
- Hall TA (1999) BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41: 95-98.
- Drummond AH, Lemey JP, Oliveira De, Pybus T, Shapiro O, et al. (2006) Tree Figure Drawing Tool Version 1.4.2. Andrew Rambaut Institute of Evolutionary Biology, University of Edinburgh, Scotland, UK.
- Sali A, Blundell TL (1993) Comparative protein structure modeling by satisfaction of spatial restraints. J MolBiol 234: 779-815.
- Discovery Studio Visualizer (2017) V41014169 2005-14 ASI.
- Nei MKS (2000) Molecular evolution and phylogenetics. Oxford University Press, New York.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30: 2725-2729.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences