Identification of Genetic Diseases Using Breast Milk Cell Analysis: The Case of Transient Neonatal Zinc Deficiency (TNZD)
Yarden Golan, Baruch Yerushalmi, Edna Efrati, Yehuda G Assaraf
DOI10.21767/2573-5365.100031
Yarden Golan1, Baruch Yerushalmi2, Edna Efrati3 and Yehuda G Assaraf1*
1The Fred Wyszkowski Cancer Research Laboratory, Department of Biology, Technion-Israel Institute of Technology, Haifa, Israel
2Pediatric Gastroenterology Unit, Soroka University Medical Center and Faculty of Health Sciences, Ben-Gurion University of the Negev, Beer-Sheva, Israel
3Laboratory of Toxicology, Pharmacology and Pharmacogenetics, Rambam Health Care Campus, Haifa, Israel
- *Corresponding Author:
- Yehuda G Assaraf
The Fred Wyszkowski Cancer Research Laboratory, Department of Biology, Technion-Israel Institute of Technology, Haifa 32000, Israel.
Tel: 97248294211
Fax: 97248225153
E-mail: assaraf@technion.ac.il
Received date: April 28, 2017; Accepted date: May 18, 2017; Published date: May 23, 2017
Citation: Golan Y, Yerushalmi B, Efrati E, Assaraf YG (2017) Identification of Genetic Diseases Using Breast Milk Cell Analysis: The Case of Transient Neonatal Zinc Deficiency (TNZD). Cell Mol Med 3(2): 8.
Abstract
Infants exclusively breastfed with zinc-deficient milk, develop transient neonatal zinc deficiency (TNZD) due to inactivating mutations in the maternal zinc transporter SLC30A2/ZnT2. In the current study, DNA sequencing of SLC30A2/ ZnT2 in a mother exclusively breastfeeding her infant which presented with TNZD symptoms, identified a single nucleotide missense mutation c.839G>T. This mutation was initially predicted to result in a single amino acid substitution p.(Gly280Val). However, c.838G was also predicted to be located close to a splice site. Hence, using sequencing of mRNA isolated from the mother’s breast milk cells, we discovered that the c.839G>T mutation creates an alternative 3’-splice sites, introducing a 27 bp exon skipping. This mutant protein lacking 9 amino acids (Δ9AA) failed to dimerize with the wild type (WT) ZnT2 protein and displayed enhanced degradation. Furthermore, the zinc transport ability of this mutant protein was completely lost as shown by intracellular zinc accumulation using the zinc fluorescent probes Zinquin and Fluozin 3 AM. These findings reveal that the Δ9AA-ZnT2 variant did not exert a dominant negative effect over the WT ZnT2 protein. Hence, we provide direct evidence that a haploinsufficiency state occurs in women with heterozygous SLC30A2/ZnT2 mutations. Importantly, we herein developed a novel genetic test of expressed genes using maternal breast milk cells, which is well suited for the early diagnosis and prevention of TNZD and possibly other micronutrient deficiencies.
Keywords
Zinc transporter; Neonatal zinc deficiency; Breast milk; Early diagnosis; Haploinsufficiency
Introduction
Cumulative evidence indicates that some exclusively breastfed infants suffer from severe zinc deficiency due to lack of zinc in the breast milk they consume [1-8]. The main initial symptoms of zinc deficiency are dermatitis, diarrhea, alopecia, and loss of appetite [9]. However, some cases of zinc deficiency are misdiagnosed and infants are treated with drugs against eczema or impetigo. Infants are particularly vulnerable to zinc deficiency as they require large amount of zinc for their normal growth and development. In this respect, preterm infants are born with low hepatic zinc stores and display a low intestinal zinc absorption capacity [10]; hence, they are more prone to zinc deficiency in the first months of life [11].
Two distinct genetic disorders can cause zinc deficiency in infants. Acrodermatitis Enteropathica (AEZ; OMIM #201100) Identification of Genetic Diseases Using Breast Milk Cell Analysis: The Case of Transient Neonatal Zinc Deficiency (TNZD) Abstract Infants exclusively breastfed with zinc-deficient milk, develop transient neonatal zinc deficiency (TNZD) due to inactivating mutations in the maternal zinc transporter SLC30A2/ZnT2. In the current study, DNA sequencing of SLC30A2/ ZnT2 in a mother exclusively breastfeeding her infant which presented with TNZD symptoms, identified a single nucleotide missense mutation c.839G>T. This mutation was initially predicted to result in a single amino acid substitution p.(Gly280Val). However, c.838G was also predicted to be located close to a splice site. Hence, using sequencing of mRNA isolated from the mother’s breast milk cells, we discovered that the c.839G>T mutation creates an alternative 3’-splice sites, introducing a 27 bp exon skipping. This mutant protein lacking 9 amino acids (Δ9AA) failed to dimerize with the wild type (WT) ZnT2 protein and displayed enhanced degradation. Furthermore, the zinc transport ability of this mutant protein was completely lost as shown by intracellular zinc accumulation using the zinc fluorescent probes Zinquin and Fluozin 3 AM. These findings reveal that the Δ9AA-ZnT2 variant did not exert a dominant negative effect over the WT ZnT2 protein. Hence, we provide direct evidence that a haploinsufficiency state occurs in women with heterozygous SLC30A2/ZnT2 mutations. Importantly, we herein developed a novel genetic test of expressed genes using maternal breast milk cells, which is well suited for the early diagnosis and prevention of TNZD and possibly other micronutrient deficiencies. Keywords: Zinc transporter; Neonatal zinc deficiency; Breast milk; Early diagnosis; Haploinsufficiency Received: April 28, 2017; Accepted: May 18, 2017; Published: May 23, 2017 is an autosomal recessive disease [12,13] which presents after weaning. Acrodermatitis Enteropathica is a result of loss of function mutations in the zinc transporter Zip4 (SLC39A4), which leads to impaired intestinal absorption of zinc [14]. Hence, infants harboring Acrodermatitis Enteropathica must receive zinc supplementation for their whole life. In contrast, transient neonatal zinc deficiency (TNZD; OMIM #608118) appears only in exclusively breastfed infants as a result of low zinc concentration in the breast milk of their nursing mothers [9]. Thus, infants harboring TNZD have no zinc absorption impairment and are completely cured after zinc supplementation of their breast milk and after weaning. TNZD is predominantly a result of loss of function mutations in the SLC30A2 gene [1,3-5,7,8,15]. SLC30A2 encodes for the zinc transporter ZnT2 which is responsible for zinc accumulation in intracellular vesicles, in lactating mammary epithelial gland cells. Through exocytosis, these zincloaded vesicles secrete zinc into breast milk [16]. Excluding one case of compound ZnT2 mutations [4], and two cases with no detectable mutations in the coding region of SLC30A2 (although the promoter region was not sequenced) [17,18], all TNZD cases reported to date were found in mothers harboring heterozygous loss of function mutations in SLC30A2.
Breast milk zinc concentrations vary largely within and between mothers at different stages of lactation [19]. As a result, it is difficult to determine the reference levels of zinc concentration in human breast milk [19]. However, it appears that in cases of TNZD, zinc levels in breast milk are much lower compared to control breast milk determinations from the first days of lactation [2]. Taking into consideration the large variability in zinc concentrations in breast milk, and the fact that there is no gold standard method and an established protocol for reliable quantification of zinc concentration in breast milk, it remains highly essential to develop a reliable, easy, rapid, and noninvasive tool for early diagnosis of TNZD.
Towards this end, we herein report on the development of a novel genetic approach for the early diagnosis of ZnT2 mutations associated with TNZD using genetic analysis of mRNA isolated from breast milk cells. Specifically, we identified and characterized a novel ZnT2 mutant protein which results from impaired splicing with consequently decreased ZnT2 protein stability as well as disruption of ZnT2 homodimerization. It should be emphasized that this impaired splicing could not be predicted from the genomic single nucleotide mutation in the SLC30A2 gene. This TNZD case is the first step towards the early genetic diagnosis of other genetic disorders which are associated with proteins that are expressed in mammary epithelial gland cells (Figure 1).
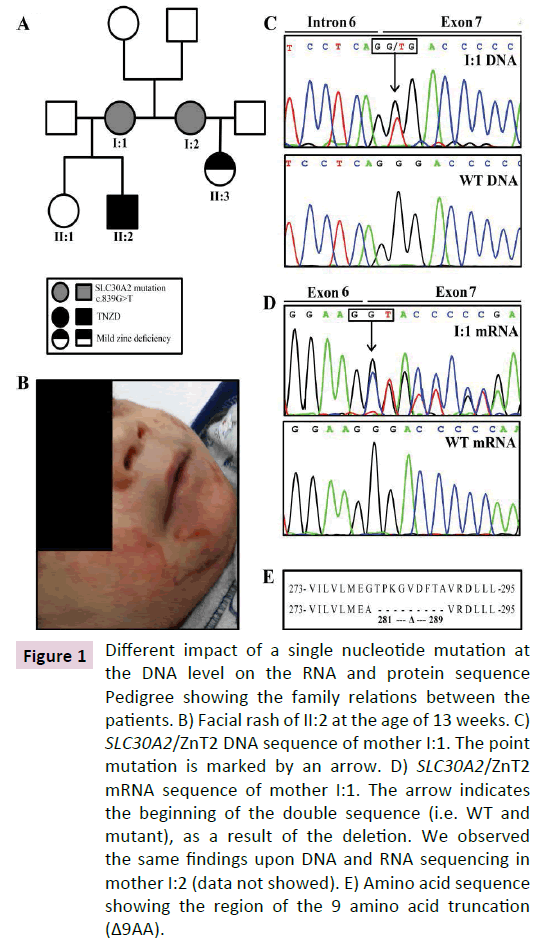
Figure 1 Different impact of a single nucleotide mutation at the DNA level on the RNA and protein sequence Pedigree showing the family relations between the patients. B) Facial rash of II:2 at the age of 13 weeks. C) SLC30A2/ZnT2 DNA sequence of mother I:1. The point mutation is marked by an arrow. D) SLC30A2/ZnT2 mRNA sequence of mother I:1. The arrow indicates the beginning of the double sequence (i.e. WT and mutant), as a result of the deletion. We observed the same findings upon DNA and RNA sequencing in mother I:2 (data not showed). E) Amino acid sequence showing the region of the 9 amino acid truncation (Δ9AA).
Materials and Methods
Chemicals and reagents
The DNA dye Hoechst 33342 was purchased from Sigma-Aldrich. The cell permeant, viable fluorescent zinc probe Zinquin ethyl ester was obtained from Biotium (Hayward, CA), whereas Fluozin3-AM was from Thermo Fisher Scientific. Zinc sulfate was obtained from Merck.
Determination of zinc concentration in breast milk
Fresh human milk samples were collected from mothers and were stored at -20°C until analysis. Zinc quantification was performed using electrothermal atomic absorption spectrometry as described previously [20] . Each sample was quantified in duplicates on two different days. Genome Sequence Analysis of SLC30A2-The current study was approved by the Institutional Review Board of the RB Rappaport Faculty of Medicine (US-HHSFWA- 00013345, 20-2016). Written consent was obtained from all subjects. Blood samples were derived from two women with low zinc milk concentration and their available family members. DNA was extracted from blood using DNeasy Blood and Tissue Kit (Qiagen). Oligonucleotide primers were designed to amplify each of the 8 exons of ZnT2 as described previously [21]. PCR products were isolated using PCR Clean-up system (Promega Corporation, WI, USA) and sequenced by Hy-labs, Israel (Table 1).
| Nucleotide substitution | Predicted amino acids substitution | Actual impact on the ZnT2 protein | Reference |
|---|---|---|---|
| c.838G > A | p.Gly280Arg | Unknown | [3, 5] |
| c.839G > T | p.Gly280Val | Exon skipping of 27 bp, caused a deletion of 9 amino acids. Gly280 was substituted by Alanine and following 8 amino acids (p.Thr281_Ala289) were deleted. | Current paper |
Table 1 Actual impact of single nucleotide substitution on the ZnT2 protein.
Sequence analysis of SLC30A2 mRNA
Fresh human milk was collected once from the 4 mothers (to sisters and to healthy individuals) in the morning hours using an electric vacuum pump and the milk was transferred to the laboratory on ice (up to 3 hours). Milk samples (30 ml) were then centrifuged at 2,000xg for 5 min at 4°C. Total RNA was extracted from the pellet containing the total cell populations, using Tri- Reagent (Sigma-Aldrich). cDNA was synthesized from total RNA (1 μg) using a high capacity cDNA reverse transcription kit (Applied Biosystems, Foster city, CA, USA). cDNA was amplified using a 5xReadyMix PCR master mix reaction buffer (AB gene, Surrey, UK; total volume 50 μl) consisting of 10 pmol of each primer according to the instructions of the manufacturer. The primers for the PCR analysis are listed in Table 2. PCR was performed using the following conditions: initial melting at 95°C for 5 min, followed by 35 cycles each of 1 min at 95°C, annealing of 1 min at 60°C, elongation of 1 min at 72°C, followed by 10 min extension at 72°C. PCR products were isolated using PCR Clean-up system (Promega Corporation, WI, USA) and sequenced by Hy-labs, Israel. Nucleotide numbering is based on the CDS of SLC30A2 mRNA variant 1 (NM_001004434.2).
| Purpose | Name | Forward | Reverse |
|---|---|---|---|
| Site-directed mutagenesis recapitulating the deletion of 27bp of ZnT2 | ZnT2-del-839-865 | GGTGTTGATGGAAGCTGTTCGTGATCTGC | GCAGATCACGAACAGCTTCCATCAACACC |
| ZnT2 mRNA sequencing | ORF1-ZnT2 | ACTGCATGGAGGCCAAGGAG | GTCGCCGATCACATGGATG |
| ZnT2 mRNA sequencing | ORF2-ZnT2 | CTGGTGTACCTGGCTGTGGAG | TGAGCAGTCAGTCTGAGGGGC |
| Real-time | Δ9AA-4mm-ZnT2 | GATCCTGGTGTTGATGGATGCT | TGAGCAGTCAGTCTGAGGGGC |
| Real-time | WT-ZnT2 | CCAAGGGCGTTGACTTCACA | GATGTGGACAGACAGAACAGGCTGG |
Table 2 Primers used for PCR.
Real-time PCR for allele expression quantification
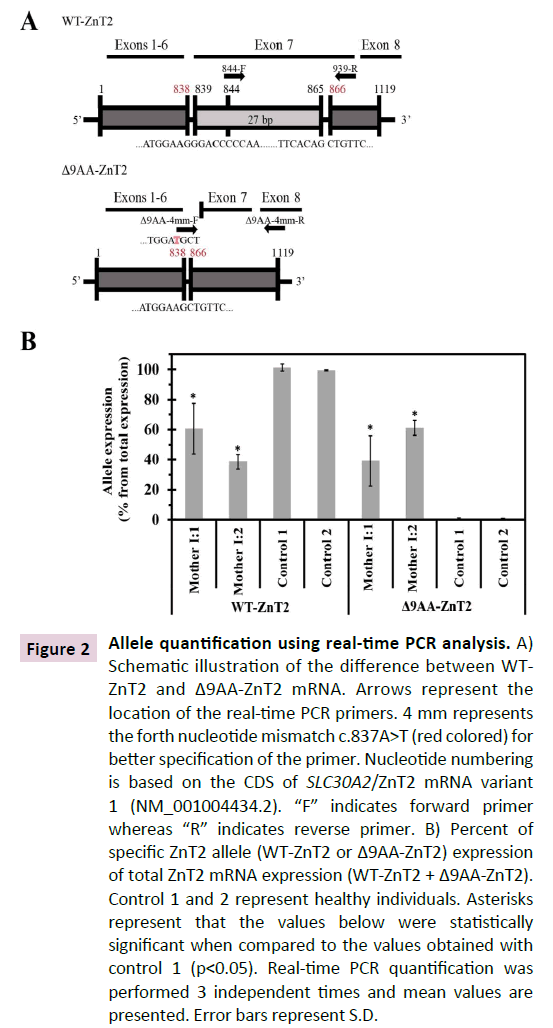
cDNA was synthesized from RNA isolated from the cell fraction derived from breast milk samples obtained from the two mothers and two control healthy individuals with matched infant ages (7 months old). Diagnostic primers were designed to detect the two different alleles i.e. the WT and the truncated Δ9AA variant. The first primer pair was designed to detect only the WT-ZnT2 transcript as the forward primer targeted the 21 bp that were deleted from the mRNA of the Δ9AA variant (Figure 2A, 844-F). In contrast, the second forward primer was designed to detect only the deleted mRNA and hybridized solely to the mRNA that lacks the 27 bp (Figure 2A- Δ9AA-4mm-F). For this purpose, we designed a forward primer which contains 19 bp that hybridized with the sequence upstream to the deleted segment, and 2 bp following the deletion (Figure 2A-Δ9AA-4mm-F). In addition, for better specification, we added a mismatch in the fourth nucleotide at the 3’ of the forward primer c.837A>T (Figure 2A-Δ9AA-4mm-F) [22]. Real-time PCR was performed using an Applied Biosystems- Life Technologies 7300 real-time PCR. A quantitative PCR reaction (20μl) contained 5ng of cDNA. PerfeCTa SYBR Green FastMix reagent was used (Quantabio, MA, USA). The primers used for allele quantification are listed in Table 2. We first performed a real-time PCR on known template amounts and compared the threshold cycle (Ct) values obtained with each primer; we found no difference between the efficacies of the two primers. For each sample we performed a triplicate quantification and then calculated the mean Ct. DNA quantification values for each primer pair in each sample were calculated using the parameters of the standard curve (b: intercept; a: slope) as follows: value=10[(Ctb)/ a] [23] . For each sample, we calculated the percentage of relative expression of each allele from the total expression (total expression was the sum of the two allele values).
Figure 2 Allele quantification using real-time PCR analysis. A) Schematic illustration of the difference between WTZnT2 and Δ9AA-ZnT2 mRNA. Arrows represent the location of the real-time PCR primers. 4 mm represents the forth nucleotide mismatch c.837A>T (red colored) for better specification of the primer. Nucleotide numbering is based on the CDS of SLC30A2/ZnT2 mRNA variant 1 (NM_001004434.2). “F” indicates forward primer whereas “R” indicates reverse primer. B) Percent of specific ZnT2 allele (WT-ZnT2 or Δ9AA-ZnT2) expression of total ZnT2 mRNA expression (WT-ZnT2 + Δ9AA-ZnT2). Control 1 and 2 represent healthy individuals. Asterisks represent that the values below were statistically significant when compared to the values obtained with control 1 (p<0.05). Real-time PCR quantification was performed 3 independent times and mean values are presented. Error bars represent S.D.
Expression vector construction
ZnT2 BiFC constructs were generated as previously described [24] . The nucleotide deletion was introduced into the ZnT2 expression plasmids using Pfu Turbo DNA polymerase (QuikChange kit, Stratagene, La Jolla, CA) and primers listed in Table 2. The coding region of the Ruby tag was amplified using mRuby- Lifeact-7 expression vector and the primers Xho-pmRuby-F 5’ATACTCGAGACCATGAACAGCCTGATCAAA 3’ and mRuby- Xba-R 5’ ATATCTAGATTACCCTCCGCCCAGGCCG 3’.
The PCR products were then sequenced to verify that they contained the correct length of the insert using agarose gel electrophoresis, following which the DNA was purified from the gel using a Promega Wizard SV gel and PCR cleanup system. The purified PCR products were digested with the appropriate restriction enzymes and were cloned into the pcDNA3.1-Zeo- ZnT2-containing constructs in a vector: insert ratio of 1:6 using the same restriction enzymes. The ligation was performed for 30 min at room temperature and the ligation products were transformed into heat shock competent E. coli DH5α. Positive colonies were selected using PCR. The fidelity of the insert and the tag were confirmed by direct sequencing (Hy-labs, Israel).
BiFC terminology
YN and YC represent the non-fluorescent N- and C-terminal halves of YFP, respectively. Transfection with the construct- YC-YN implies that plasmids containing the construct-YN and the construct-YC were co-transfected into cells. Construct-YFP indicates that plasmids containing the full-length YFP fluorescent protein were transfected.
Cell culture, transient transfections, flow cytometry and confocal laser microscopy
MCF-7 breast cancer cells were grown and transiently transfected as previously described [3,24] for all the experiments presented here. Twenty-four hrs after transfection, cells were stained with Zinquin ethyl ester (40 μM) or Fluozin3-AM (1μM) and were analyzed using flow cytometry or confocal microscopy, respectively. For the zinc transport function experiments, cells were incubated for 2 hrs with growth medium containing 75 μM ZnSO4 before staining. Cells were then rinsed with PBS and incubated in growth medium containing 40 μM Zinquin ethyl ester and were analyzed with a flow cytometer as previously described [3,25] . For confocal microscopy, cells were incubated in growth medium containing 1 μM Fluozin3-AM for 30 min, following which they were washed 3 times with PBS. Cells were then incubated for another 30 min in growth medium at 37°C to allow a complete de-esterification of intracellular AM esters. Finally, cells were washed with PBS and then incubated in PBS containing 1 mM MgCl2, 1 mM CaCl2 and 10 mM D-glucose at pH 7.4. Hoechst 33342 (2 μg/ml) was used for viable nuclear DNA staining. Live cells were imaged using an inverted confocal microscope (Zeiss LSM 710) at a magnification of X63 under immersion oil.
Assessment of ZnT2 dimerization in BiFC transfectants using Western blot analysis
MCF-7 cells were transiently transfected with the designated plasmids (3 μg in single construct transfection and 1.5 μg DNA of each construct in co-transfections). Membrane proteins were isolated on ice using an extraction buffer as previously described [25] . Briefly, proteins (30 μg) were resolved by electrophoresis on 10% polyacrylamide gels, electroblotted onto cellulose nitrate membranes, blocked with Tris-buffered saline (TBS)-milk and reacted with mouse anti-ZnT2 monoclonal antibody (at 1:2,000 dilutions, 1 hr in room temperature; generously provided by Prof. T. Kambe, Kyoto University, Kyoto, Japan). Blots were then washed three times with TBS for 10 min each at room temperature and incubated with horseradish peroxidase-conjugated goat antimouse IgG (1:20,000 dilution; Jackson Immunoresearch Labs, West Grove, PA; catalog number: 115035062) for 1 hr at room temperature. After three washes, enhanced chemiluminescence detection was performed according to the manufacturer's instructions (Biological Industries, Beth-Haemek, Israel). Similarly, actual protein loading onto the SDS-PAGE gels was confirmed by blot reprobing with a rabbit polyclonal antibody against the α subunit of Na+/K+ ATPase (KETTY at 1:3,000 dilutions; kindly provided by Prof. S.J.D. Karlish, Weizmann Institute of Science, Rehovot, Israel) and detected with horseradish peroxidaseconjugated goat anti-rabbit IgG (1:15,000 dilutions; Jackson Immuno-Research Labs, West Grove, PA; catalog number: 115035045).
Detection of native human ZnT2
Breast milk (30 ml) was centrifuged at 2,000xg for 5 min at 4°C and was separated into 3 fractions. Membrane protein extraction buffer [24] (100 μl) was added directly to the fraction of cells (bottom fraction). Samples were then incubated on ice for 30 min and centrifuged at 20,000xg for 15 min at 4°C. The fraction containing membrane proteins was collected and proteins (20 μg) resolved by electrophoresis on 10% polyacrylamide gels as described above. Mouse anti-ZnT2 monoclonal antibody was used for Western blot analysis (at 1:3,000 dilution, 1 hr. in room temperature). Actual protein loading onto the SDS-PAGE gels was confirmed by blot reprobing with a rabbit polyclonal antibody against actin (at 1:1,000 dilution, O.N. at 4°C; Sigma-Aldrich; catalog number: A2066).
Evaluation of the half-life of WT and mutant ZnT2 using cycloheximide inhibition of translation
MCF-7 cells were seeded and transiently transfected as described above. Sixteen hrs. after transfection, the growth medium was replaced by fresh medium containing 50 μg/ml cycloheximide (CHX). Membrane proteins were isolated every 0.5 to 1 hr using an extraction buffer and Western blot analysis was performed as described above.
Quantification of protein band intensity after Western blot analysis
Western blot analysis was preformed using Quantity One® 1-D analysis software, by Bio-Rad Laboratories, Inc.
Statistical Analysis
Results are presented as means ± S.D. Statistical comparisons were performed using two tailed Student’s t test (Prism Graph Pad, Berkeley, CA), and a significant difference was demonstrated when p value was <0.05. Results from at least three independent experiments are shown.
Results
Clinical data and early genetic diagnosis of TNZD
Preterm infant (Son 1 in Figure 1A) suffered from a severe erythematous scaly rash involving his face, neck, diaper area at the age of 13 weeks (Figure 1B). The rash was first diagnosed as recalcitrant seborrheic dermatitis with possible secondary impetigo. Therefore, the infant was treated with different ointments including topical antibiotics and steroids without improvement. After 3 weeks with worsening in the skin findings, physicians considered the possibility of zinc deficiency. Indeed, the infant’s plasma zinc levels were extremely low (4.5 μg/dL; normal range 55 μg/dL to 165 μg/dL). Zinc supplementation (5 mg/day) was initiated and within 4-5 days of supplementation, the infant’s appetite improved, weight gain was restored and the skin rash has completely vanished in 2 weeks.
We hence introduced here a new genetic screening technique based on sequencing of ZnT2 mRNA isolated from breast milk cells; we specifically sequenced the ZnT2 mRNA of Mother 1 (I:1 Figure 1A) and her sister Mother 2 (I:2 in Figure 1A) that also gave birth, and had a 4 months old baby girl without any symptoms of zinc deficiency. After identifying the very same alternative 3’-splice site variants in mRNA extracted from their breast milk cells, we offered Mother I:2 to determine zinc blood levels in her exclusively breastfed daughter (II:3 in Figure 1A). Indeed, her daughter (II:3 in Figure 1A) displayed a mild zinc deficiency (41 μg/dL, normal range 55 μg/dL to 165 μg/dL). Thus, the present study constitutes the very first case of early diagnosis of TNZD reported to date. It is important to emphasize that II:1 was also exclusively breastfed; however, mother I:1 did not notice any unusual symptoms in her daughter, as the infant had only a mild zinc deficiency which was not associated with apparent symptoms, just like her cousin (II:3 in Figure 1A), that was completely cured after weaning. Breast milk zinc concentrations were 6 μg/dL and 11 μg/dL for I:1 and I:2, respectively (normal median range 85 μg/dL at 7 months of lactation, minimum level 44 μg/dL and maximum level 136 μg/dL) [19].
DNA and RNA sequencing
DNA sequencing of I:1 and I:2 identified a single nucleotide shift resulting in a synonymous mutation c.546G>A (Nucleotide number 1 refers to the first nucleotide of the ATG codon of the long ZnT2 mRNA variant NM_001004434.2) which does not alter the protein sequence (p. Ser182=). Furthermore, this genomic DNA sequencing identified a single nucleotide missense mutation c.839G>T (Figure 1C). This missense mutation was initially predicted to result in a single amino acid substitution p.(Gly280Val) (Table 1). However, c.838G in ZnT2 was recently predicted to be located close to splice site [5]. Therefore, we undertook further experiments to explore the impact of this genomic c.839G>T mutation on the splicing of ZnT2 mRNA. Indeed, sequencing of mRNA isolated from breast milk cells revealed that the missense genomic mutation c.839G>T results in an alternative 3’ splicing, culminating in an exon skipping of 27 bp (c.839_865del). As shown in Figure 1D, starting with nucleotide c.839G, mRNA sequence analysis revealed two distinct mRNAs: a WT mRNA and a 27bp truncated transcript resulting from skipping of a 5’-region of exon 7. We next performed TA cloning of the PCR products and sequenced the insert-harboring plasmids to verify the exact nucleotide sequence of the truncated ZnT2 allele. The 27 bp skipping resulted in a substitution of p.(Gly280Ala) along with a deletion of 9 amino acids encompassing p. Thr281_Ala289del (Figure 1E). We therefore termed the 9-amino acid truncation of this exon skipped ZnT2 variant, Δ9AA. These results show that both the WT and the Δ9AA variant alleles are expressed and highlight the importance of performing a gene expression screen at the mRNA level upon identification of a genomic SLC30A2/ ZnT2 mutation.
Development of diagnostic primers for the detection of both the 27 bp truncated allele and the WT allele
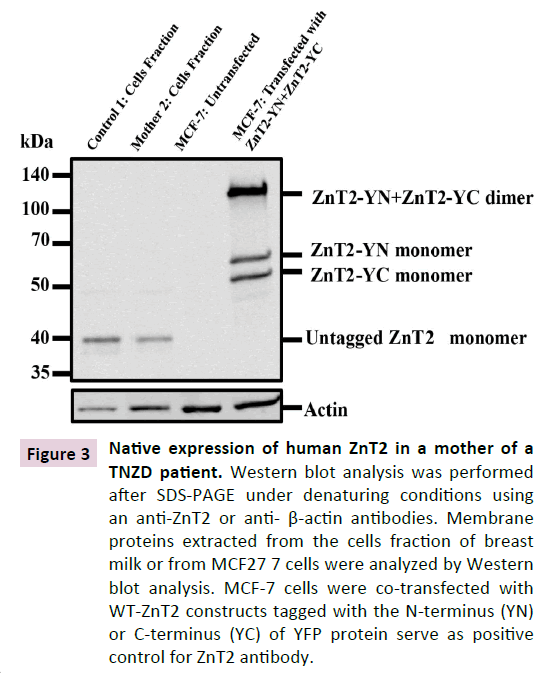
We next assessed whether or not the WT and truncated alleles are equally distributed in order to understand the molecular mechanism underlying the presumed haploinsufficiency state that we have recently proposed in TNZD [3] . To quantify the distribution of the WT and the truncated ZnT2 mRNA alleles, we designed diagnostic primers. The first set of primers was designed to detect solely the WT-ZnT2 allele (Figure 2A). In contrast, the second forward primer was selected to detect only the truncated ZnT2 mRNA (Δ9AA), hence hybridizing solely to the mRNA lacking the 27 bp (Figure 2A and Materials and Methods). We found that while both healthy individuals (Control 1,2 Figure 2B) expressed only the WT-ZnT2 allele, the two mothers harboring TNZD-associated mutations, expressed between 40% to 60% of the WT allele and the remaining transcript represented the truncated Δ9AA allele (Mother I:1,2 Figure 2B). These results demonstrate that the two alleles were comparably expressed in both mother I:1 and mother I:2. A native human ZnT2 variant (40 kDa) is expressed in breast milk cells. To verify the expression of the WT native ZnT2 protein in women with zinc-deficient breast milk, we extracted membrane proteins from the breast milk cell fraction; we found that both mother 2 and control 1 expressed predominantly the 40 kDa variant of the WT ZnT2 (Figure 3). However, taking into consideration the slight differences in the actin protein levels, one can suggest that mother 2 expressed less ZnT2 protein compared to control 1. The 9-amino acid deletion in the ZnT2 protein could not be detected on SDS-PAGE, and we therefore could not determine whether the Δ9AA variant protein was expressed. However, these findings lend further support to our suggestion that a haploinsufficiency state occurs for the unaffected WT allele in producing zinc-deficient breast milk. Although mother 2 had sufficient levels of the ZnT2 protein, her breast milk was zinc-deficient and led to a mild zinc deficiency in her infant.
Figure 3 Native expression of human ZnT2 in a mother of a TNZD patient. Western blot analysis was performed after SDS-PAGE under denaturing conditions using an anti-ZnT2 or anti- β-actin antibodies. Membrane proteins extracted from the cells fraction of breast milk or from MCF27 7 cells were analyzed by Western blot analysis. MCF-7 cells were co-transfected with WT-ZnT2 constructs tagged with the N-terminus (YN) or C-terminus (YC) of YFP protein serve as positive control for ZnT2 antibody.
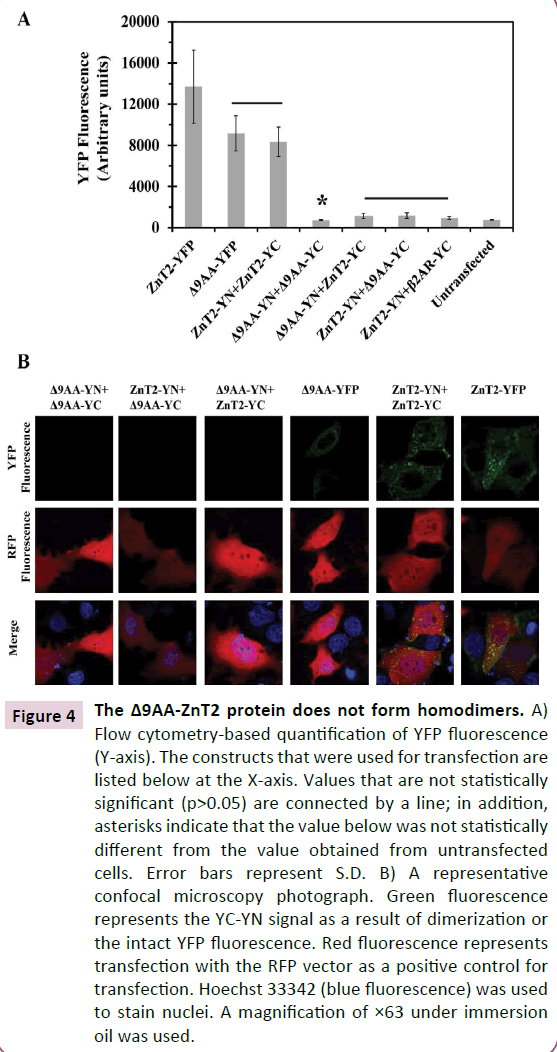
The alternative 3’ splice variant (Δ9AA) associated with TNZD, disrupts ZnT2 homodimerization. We first evaluated the ability of the alternative 3’ splice variant (Δ9AA) ZnT2 to form homodimers using the BiFC assay that we have previously applied for the detection of in situ dimerization of ZnT2 and other ZnTs in living cells [3,24,25] . High YFP fluorescence intensity indicates a strong interaction between the two tagged proteins, which enables the refolding of the two non-fluorescent halves of YFP (YN and YC) into a fully fluorescent protein. As determined by flow cytometry (Figure 4A), we found that the alternative 3’ splice ZnT2 variant which lacks 9 amino acids in the C-terminus, lost the ability to form homodimers (Figure 4A, Δ9AA -YN+Δ9AA-YC). We further found that Δ9AA -ZnT2 failed to form homodimers with the native WT-ZnT2 (Figure 4A, ZnT2-YN+ Δ9AA -YC and Δ9AA -YN+ZnT2- YC). The YFP fluorescence that we observed in all transfections with the truncated Δ9AA -ZnT2 construct was at the background level found with the negative control of the WT-ZnT2-YN+β2ARYC co-transfection. Upon transfection of the Δ9AA-YFP construct we observed relatively prominent level of YFP fluorescence which confirmed the expression of the Δ9AA-ZnT2 protein. However, the YFP fluorescence levels of the Δ9AA-YFP protein were lower compared to WT-ZnT2-YFP levels, indicating that the deletion of this 9-amino acid segment at the C-terminus possibly decreases the expression and/or stability of this truncated ZnT2 protein. This hypothesis was first evaluated using confocal laser microscopy (Figure 4B). Indeed, YFP fluorescence was neither detected upon co-transfection with the Δ9AA-YN+Δ9AA-YC constructs nor upon co-transfection with Δ9AA-YC+ZnT2-YN, or with the Δ9AAYN+ ZnT2-YC constructs (Figure 4B). These findings indicate that the truncated Δ9AA-ZnT2 protein has lost its homodimerization capacity. These results further show that the truncated Δ9AAZnT2 protein cannot exert a dominant negative effect over the WT-ZnT2 protein as it cannot form dimers.
Figure 4 The Δ9AA-ZnT2 protein does not form homodimers. A) Flow cytometry-based quantification of YFP fluorescence (Y-axis). The constructs that were used for transfection are listed below at the X-axis. Values that are not statistically significant (p>0.05) are connected by a line; in addition, asterisks indicate that the value below was not statistically different from the value obtained from untransfected cells. Error bars represent S.D. B) A representative confocal microscopy photograph. Green fluorescence represents the YC-YN signal as a result of dimerization or the intact YFP fluorescence. Red fluorescence represents transfection with the RFP vector as a positive control for transfection. Hoechst 33342 (blue fluorescence) was used to stain nuclei. A magnification of ×63 under immersion oil was used.
Δ9AA-ZnT2 protein undergoes enhanced degradation
As the above flow cytometry results with the YFP tag suggested enhanced degradation of the Δ9AA-ZnT2 protein, we first undertook experiments to explore its ability to form homodimers. As we previously showed [25], upon dimerization of WT ZnT2- YN+ZnT2-YC, the BiFC tags refold and tightly interact as a result of the formation of multiple hydrogen bonds, thereby forming dimers that are metastable in SDS-PAGE (Figure 5A). Upon transfection with Δ9AA-YN or Δ9AA-YC constructs, we observed lower ZnT2 levels, compared to transfection with the WT-ZnT2 as revealed by Western blot analysis (Figure 5A). However, in contrast to the ZnT2 protein which was predominantly present in the homodimer form (Figure 5A, upper band), upon cotransfection with Δ9AA-YN+Δ9AA-YC, no homodimers were detected (Figure 5A). Furthermore, upon co-transfection of ZnT2- YN along with Δ9AA-YC, the protein levels were substantial, yet no homodimers were detectable (Figure 5A, right side). These results indicate that the lower expression levels or increased degradation of the truncated Δ9AA ZnT2 protein were not the only causes for the loss of the dimerization capacity. These findings suggest that the deleted C-terminal region of ZnT2 has a major role in protein-protein interactions and dimer formation.
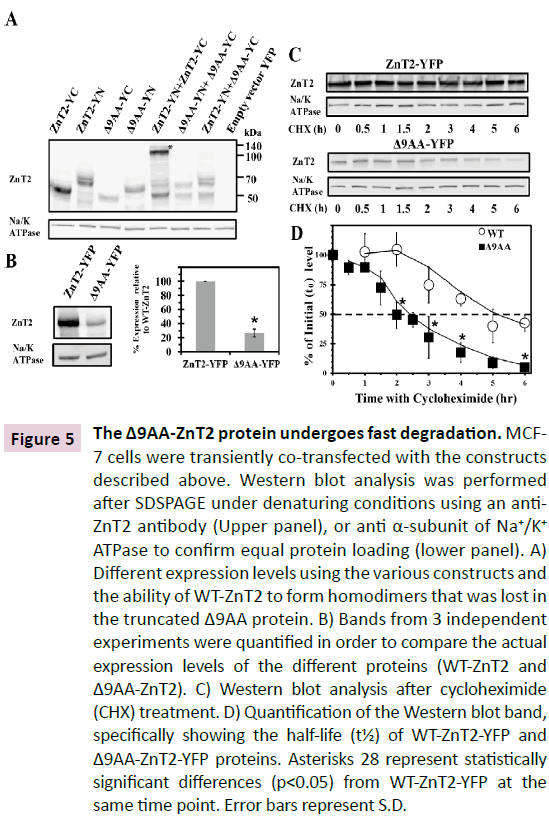
Figure 5 The Δ9AA-ZnT2 protein undergoes fast degradation. MCF- 7 cells were transiently co-transfected with the constructs described above. Western blot analysis was performed after SDSPAGE under denaturing conditions using an anti- ZnT2 antibody (Upper panel), or anti α-subunit of Na+/K+ ATPase to confirm equal protein loading (lower panel). A) Different expression levels using the various constructs and
the ability of WT-ZnT2 to form homodimers that was lost in the truncated Δ9AA protein. B) Bands from 3 independent experiments were quantified in order to compare the actual expression levels of the different proteins (WT-ZnT2 and Δ9AA-ZnT2). C) Western blot analysis after cycloheximide (CHX) treatment. D) Quantification of the Western blot band, specifically showing the half-life (t½) of WT-ZnT2-YFP and Δ9AA-ZnT2-YFP proteins. Asterisks 28 represent statistically significant differences (p<0.05) from WT-ZnT2-YFP at the same time point. Error bars represent S.D.
We next determined the protein levels of WT-ZnT2-YFP compared to the truncated Δ9AA-ZnT2-YFP (Figure 5B) after transient transfection and Western blot analysis. We found that 24 hrs after transfection, the Δ9AA-ZnT2 protein was expressed at a level that was ~25% that of the WT protein (Figure 5B). Thus, in order to determine the molecular basis underlying this decreased levels of the Δ9AA-ZnT2 protein, we directly determined the half-lives of the truncated Δ9AA-ZnT2 and the WT ZnT2 proteins using the cycloheximide (CHX) treatment method [4] . As shown in Figure 5B, we found that the levels of the truncated Δ9AA-ZnT2 protein were significantly lower at t0 compared to the WT ZnT2 protein, and more importantly, the degradation rate of the Δ9AA-ZnT2 protein was significantly enhanced when compared to that of the WT ZnT2 protein (Figure 5C). Quantification of the ZnT2 bands upon Western blot analysis revealed that the half-life (t½) of the WT-ZnT2-YFP protein was ≈4.5 hrs, whereas that of the truncated Δ9AA-ZnT2-YFP protein was only ≈2 hrs (Figure 5D). These results provide direct evidence for the enhanced degradation of the Δ9AA-ZnT2 protein, hence providing the molecular basis for the low expression levels observed with the truncated Δ9AA-ZnT2 protein, when compared to the WT-ZnT2 protein.
The alternative 3’ splice variant Δ9AA ZnT2 is devoid of zinc transport function
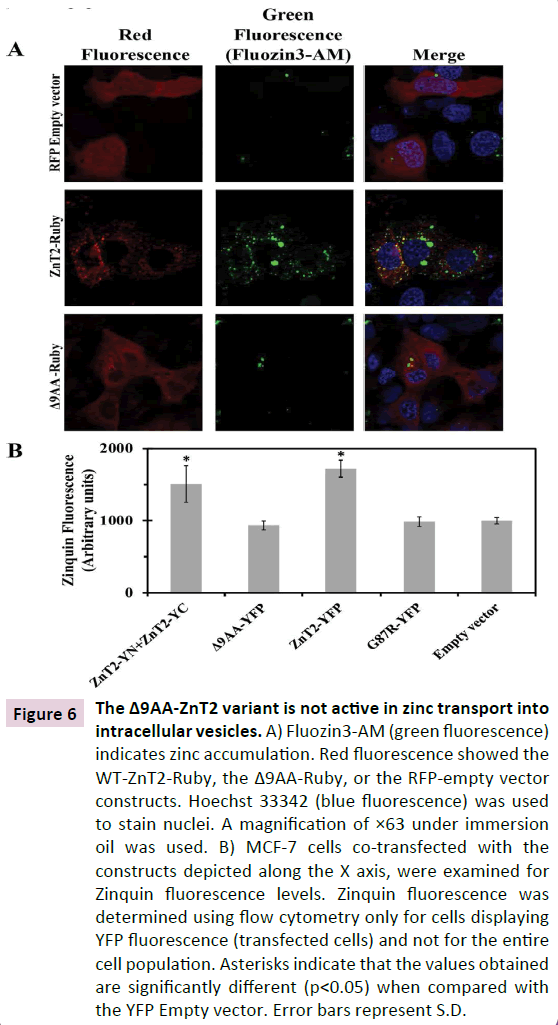
In order to determine whether or not the truncated Δ9AA ZnT2 protein retained zinc transport activity, we used two distinct viable fluorescent zinc probes, Fluozin-3 AM and Zinquin ethylester. MCF-7 cells transfected with the tagged WT-ZnT2-Ruby exhibited a high number of fluorescent intracellular vesicles upon staining with Fluozin-3, a cell-permanent fluorescent zinc indicator. This confirmed the key role that the WT ZnT2 plays in compartmentalization of zinc in intracellular vesicles. In contrast, upon transfection with the truncated Δ9AA-Ruby, we observed low levels of red fluorescence, consistent with the fast degradation of the Δ9AA-Ruby protein (Figure 6), and we also observed a very low number of intracellular zinc-containing vesicles (Figure 6A); this background number of Fluozin-3-stained vesicles in the Δ9AA-Ruby transfected cells was similar to that observed upon transfection with the RFP-Empty vector (Figure 6A). The second fluorescent zinc probe that we used was Zinquin ethyl-ester which we and others previously employed in order to follow zinc accumulation in intracellular vesicles [24,25] . We show here that in contrast to the zinc transport activity of WT-ZnT2- YFP and WT-ZnT2-YC-YN, the truncated Δ9AA-YFP protein failed to mediate the accumulation of zinc in intracellular vesicles. We therefore obtained low levels of Zinquin fluorescence that were similar to the background levels observed upon transfection with YFP empty vector (which did not contain a ZnT2 insert) or to the dominant negative p.Gly87Arg ZnT2 mutant that we previously showed to be devoid of zinc transport activity (Figure 6B). These results indicate that the Δ9AA-ZnT2 variant lost the ability to transport zinc into intracellular vesicles.
Figure 6 The Δ9AA-ZnT2 variant is not active in zinc transport into intracellular vesicles. A) Fluozin3-AM (green fluorescence) indicates zinc accumulation. Red fluorescence showed the WT-ZnT2-Ruby, the 9AA-Ruby, or the RFP-empty vector constructs. Hoechst 33342 (blue fluorescence) was used to stain nuclei. A magnification of ×63 under immersion oil was used. B) MCF-7 cells co-transfected with the constructs depicted along the X axis, were examined for Zinquin fluorescence levels. Zinquin fluorescence was determined using flow cytometry only for cells displaying YFP fluorescence (transfected cells) and not for the entire
cell population. Asterisks indicate that the values obtained are significantly different (p<0.05) when compared with the YFP Empty vector. Error bars represent S.D.
Discussion
Extraction of RNA from breast milk was recently shown to be a rapid, facile, and non-invasive technique for studying the gene expression profile in mammary gland epithelial cells. For example, Sharp et al. reported on the differences in the transcriptomics profiles at the various stages of lactation [23]. Furthermore, C. Cai and his group studied the role of the gene expression profile of mammary gland epithelial cells isolated from breast milk in the iron uptake pathway [26] . Hence, we herein developed a novel genetic tool for the identification of distinct ZnT2 mutations and genetic variants using cells isolated from breast milk at the expressed mRNA level. This genetic analysis can serve as a reliable screening platform for the early diagnosis of TNZD and can prevent zinc deficiency in infants before the severe symptoms appear. In the case of TNZD, the early diagnosis using breast milk cells is critical because of the following reasons:
The diagnosis of mild zinc deficiency is very complex, and most cases remain undiagnosed or misdiagnosed until the severe symptoms emerge. In addition, the zinc level blood test is not properly reflecting the exact body zinc status, at the middle ranges i.e. between normal levels and severe deficiency [27] .
Infants at the age of one month to one year are not tested routinely for blood micronutrient levels, unless they have health problems.
Upon early diagnosis of TNZD, the infant can be easily supplemented with zinc and can continue breastfeeding without any pathological symptoms.
We showed here for the first time, that single nucleotide mutations at the genomic DNA level in the ZnT2 gene may result in completely distinct deleterious alterations at the mRNA and amino acids levels which could not be simply predicted from the genomic mutation identified. In this respect, alterations at the genomic DNA level in a given genetic disease may predict some impact on the protein level which may be completely erroneous and misleading. In contrast, at the expressed mRNA level, the deleterious impact of the mutation will be most accurate and fully representative. Consistently, we herein successfully detected this unique ZnT2 splice variant only when using cells isolated from breast milk as ZnT2 is not ubiquitously expressed in different tissues.
We previously suggested a haploinsufficiency state for the unaffected WT ZnT2 allele in TNZD pathology [3]. This suggestion was based on three different heterozygous mutations that caused TNZD without exerting a dominant negative effect over the WT ZnT2 protein [3,5]. In the current paper, we provide direct support for this hypothesis as shown in the TNZD case we characterized here. Specifically, while the WT allele transcript was nearly equally expressed as the mutant allele (Figure 2), and there was sufficient native expression of ZnT2 protein (Figure 3), the truncated Δ9AA-ZnT2 protein still failed to form homodimers with the WT-ZnT2 protein (Figure 4). This was true in spite of the fact that the actual cellular WT-ZnT2 protein levels were significantly higher than those of the Δ9AA-ZnT2 protein (Figure 5). Hence, these high levels of the WT-ZnT2 protein were not sufficient to mediate adequate zinc transport to meet the infant’s requirements (Figure 1, Son 1). We further suggest that production of zinc-deficient milk leading to TNZD is an autosomal dominant disease which exhibits incomplete penetrance. Other factors that may affect the appearance of TNZD in the infant are gestational age at birth, birth weight, intrinsic liver zinc stores at birth [28], milk formula consumption and more. It is important to note that Son 1 was born as a premature infant, as it is well known that premature infants are prone to zinc deficiency because of their meager hepatic zinc stores and their low intestinal zinc absorption capacity [11] . However, most premature infants that consume breast milk containing adequate zinc concentrations will not develop severe zinc deficiency. In addition, daughter 2 which was born in term (after 40 weeks of gestation), was found to harbor a mild zinc deficiency at the age of 4 months without any serious symptoms. This infant could develop a severe zinc deficiency at a later stage if she was left untreated and was continuing exclusive breastfeeding or could remain at a mild zinc deficiency state without being diagnosed. Mild zinc deficiency in children was linked to neurological and cognitive changes [29], growth retardation [30], immune deficiency and higher morbidity of diarrhea and pneumonia [27].
The nonspecific symptoms of mild zinc deficiency make it hard to diagnose and as a result, most cases remain undiagnosed and hence untreated [31]. The current study constitutes the first case of early diagnosis of zinc deficiency in an infant that was treated with zinc supplements and TNZD was prevented. Using our novel genetic screening tool with ZnT2 mRNA isolated from breast milk cells, we could easily identify a breast milk sample from mothers at the first days of lactation and prevent infant’s exclusive nursing by mothers harboring a ZnT2 mutation which inflicts the severe symptoms of TNZD. Furthermore, this novel diagnostic technique could be also applied for the early identification of other inborn genetic diseases such as riboflavin deficiency which is common in breastfed preterm infants that may be result from mutations in the ABCG2 (BCRP) transporter that is responsible for riboflavin secretion to breast milk [33,34]. In addition, using this tool we could further investigate the underlying molecular mechanism of transport of other micro- and macronutrients. Importantly, one could utilize this breast milk RNA isolation platform to detect and identify mastitis for personalized treatment depending on the origin of infection being fungal or bacterial [35].
In summary, we presented here the first case of early diagnosis of TNZD and applied a non-invasive tool for the prevention of TNZD in infants which are exclusively breastfed by mothers harboring loss of function mutations in SLC30A2/ZnT2 gene. We provide direct evidence that a haploinsufficiency state occurs in women harboring heterozygous SLC30A2/ZnT2 gene mutations, hence suggesting that production of zinc-deficient breast milk is an autosomal dominant inheritance-based phenotype. The appearance of TNZD in the nursing infant showed a partial penetrance pattern depending on other intrinsic and environmental factors. Importantly, we highlight the importance of sequencing at the expressed mRNA level compared to genomic DNA sequencing for the accurate identification of the molecular mechanism underlying distinct genetic diseases.
References
- Chowanadisai W, Lonnerdal B, Kelleher SL (2006) Identification of a mutation in SLC30A2 (ZnT-2) in women with low milk zinc concentration that results in transient neonatal zinc deficiency. J Biol Chem 281: 39699-39707.
- Glover MT, Atherton DJ (1988) Transient zinc deficiency in two full-term breast-fed siblings associated with low maternal breast milk zinc concentration Pediatr Dermatol 5: 10-13.
- Golan Y, Itsumura N, Glaser F (2016) Molecular Basis of Transient Neonatal Zinc Deficiency: NOVEL ZnT2 mutations disrupting zinc binding and permeation. J Biol Chem 291: 1354-13559.
- Itsumura N, Inamo Y, Okazaki F (2013) Compound heterozygous mutations in SLC30A2/ZnT2 results in low milk zinc concentrations: A novel mechanism for zinc deficiency in a breast-fed infant. PLoS One 8: e64045.
- Itsumura N, Kibihara Y, Fukue K (2007) Novel mutations in SLC30A2 involved in the pathogenesis of transient neonatal zinc deficiency. Pediatr Res
- Kienast A, Roth B, Bossier C (2007) Zinc-deficiency dermatitis in breast-fed infants. Eur J Pediatr 166: 189-194.
- Lova Navarro M, Vera Casano A, Benito Lopez C (2014) Transient neonatal zinc deficiency due to a new autosomal dominant mutation in gene SLC30A2 (ZnT-2). Pediatr Dermatol 31: 251-252.
- Miletta MC, Bieri A, Kernland K (2013) Transient Neonatal Zinc Deficiency Caused by a Heterozygous G87R Mutation in the Zinc Transporter ZnT-2 (SLC30A2) Gene in the Mother Highlighting the Importance of Zn (2+) for Normal Growth and Development. Int J Endocrinol 259189.
- Kasana S, Din J, Maret W (2015) Genetic causes and gene-nutrient interactions in mammalian zinc deficiencies: Acrodermatitis enteropathica and transient neonatal zinc deficiency as examples. J Trace Elem Med Biol 29: 47-62.
- Kambe T, Fukue K, Ishida R (2015) Overview of inherited zinc deficiency in infants and children. J Nutr Sci Vitaminol 61: S44-46.
- Dorea JG (2002) Zinc deficiency in nursing infants. J Am Coll Nutr 21: 84-87.
- Coromilas A, Brandling-Bennett HA, Morel KD (2011) Novel SLC39A4 mutation in acrodermatitis enteropathica. Pediatr. Dermatol 28: 697-700.
- Kambe T, Tsuji T, Hashimoto A (2015) The physiological, biochemical, and molecular roles of zinc transporters in zinc homeostasis and metabolism. Physiol Rev 95: 749-784.
- Kury S, Dreno B, Bezieau S (2002) Identification of SLC39A4, a gene involved in acrodermatitis enteropathica. Nat Genet 31: 239-240.
- Qian L, Wang B, Tang N (2012) Polymorphisms of SLC30A2 and selected perinatal factors associated with low milk zinc in Chinese breastfeeding women. Early Hum Dev 88: 663-668.
- Kasana S, Din J, Maret W (2015) Genetic causes and gene-nutrient interactions in mammalian zinc deficiencies: Acrodermatitis enteropathica and transient neonatal zinc deficiency as examples. J Trace Elem Med Biol 29: 47-62.
- Kumar L, Michalczyk A, McKay J (2015) Altered expression of two zinc transporters, SLC30A5 and SLC30A6, underlies a mammary gland disorder of reduced zinc secretion into milk. Genes Nutr 10: 487.
- Yang WL, Hsu CK, Chao SC (2012) Transient zinc deficiency syndrome in a breast-fed infant due to decreased zinc in breast milk (type II hypozincemia of infancy): A case report and review of the literature.
- Dorea JG (2000) Zinc in human milk. Nutr Res 20: 1645-1687.
- Arnaud J, Favier A, Bye AME (1992) Determination of ultra-filterable zinc in human milk by electrothermal atomic absorption spectrometry. Analyst 117: 1593.
- Lasry I, Seo YA, Ityel H (2012) A dominant negative heterozygous G87R mutation in the zinc transporter, ZnT-2 (SLC30A2), results in transient neonatal zinc deficiency. J Biol Chem 287: 29348-29361.
- Kamel AA-E, Huo N, Gu YQ (2003) Bioinformatic tools and guideline for PCR primer design. African J Biotechnol 2: 91-95.
- Hagège H, Klous P, Braem C (2007) Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat Protoc 2: 1722-1773.
- Lasry I, Golan Y, Berman B (2014) In situ dimerization of multiple wild type and mutant zinc transporters in live cells using bimolecular fluorescence complementation. J Biol Chem 289: 7275-7292.
- Golan Y, Berman B, Assaraf YG (2015) Heterodimerization, altered subcellular localization, and function of multiple zinc transporters in viable cells using bimolecular fluorescence complementation. J Biol Chem 290: 9050-9063.
- Cai C, Eck P, Friel JK (2016) Gene expression profiles suggest iron transport pathway in the lactating human Epithelial Cell. J Pediatr Gastroenterol Nutr 1.
- Hambidge M (2000) Zinc and health: Current status and future directions human zinc deficiency 1. Am Soc Nutr Sci 1344-1349.
- Butte N, Lopez-Alaracon M, Garza C (2002) Nutrient adequacy of exclusive breastfeeding for the term infant during the first six months of life. World Heal Organ 47.
- Black MM (1998) Zinc deficiency and child development. Am J Clin Nutr 68: 464S-469S.
- Nakamura T, Nishiyama S, Futagoishi-Suginohara Y (1993) Mild to moderate zinc deficiency in short children: Effect of zinc supplementation on linear growth velocity. J Pediatr 123: 65-69.
- Hambidge KM (1989) Mild zinc deficiency in human subjects, Springer London p. 281-296.
- Lucas A, Bates C (1984) Transient riboflavin depletion in preterm infants. Arch Dis Child 59: 837-841.
- Hovi L, Hekali R, Siimes MA (1979) Evidence of riboflavin depletion in breast-fed newborns and its further acceleration during treatment of hyperbilirubinemia by phototherapy. Acta Paediatr 68: 567-570.
- van Herwaarden AE, Wagenaar E, Merino G (2007) Multidrug transporter ABCG2/breast cancer resistance protein secretes riboflavin (vitamin B2) into milk. Mol Cell Biol 27: 1247-1253.
- Brenaut P, Bangera R, Bevilacqua C (2012) Validation of RNA isolated from milk fat globules to profile mammary epithelial cell expression during lactation and transcriptional response to a bacterial infection. J Dairy Sci 95: 6130-6144.
Open Access Journals
- Aquaculture & Veterinary Science
- Chemistry & Chemical Sciences
- Clinical Sciences
- Engineering
- General Science
- Genetics & Molecular Biology
- Health Care & Nursing
- Immunology & Microbiology
- Materials Science
- Mathematics & Physics
- Medical Sciences
- Neurology & Psychiatry
- Oncology & Cancer Science
- Pharmaceutical Sciences